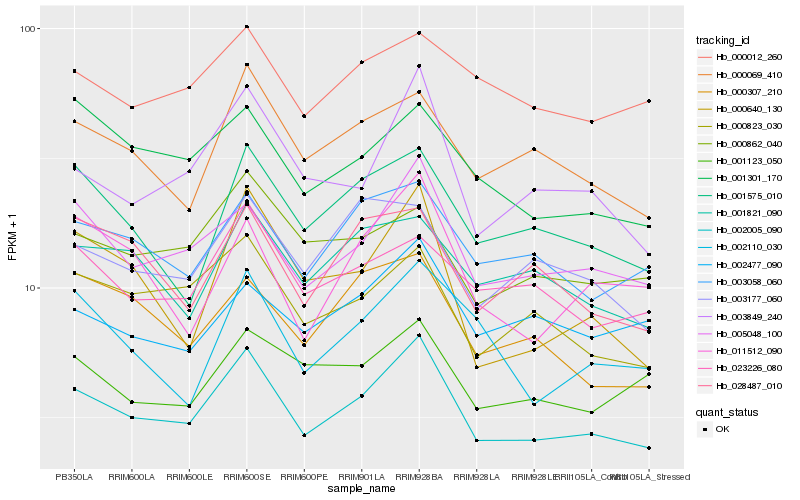
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005048_100 |
0.0670423583 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 3 |
Hb_003849_240 |
0.0823304809 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000823_030 |
0.086796241 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 5 |
Hb_000069_410 |
0.0900851447 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_003058_060 |
0.0913291491 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 7 |
Hb_001575_010 |
0.0959351145 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_023226_080 |
0.0962146709 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001301_170 |
0.0975209741 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 10 |
Hb_000862_040 |
0.0979852551 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 11 |
Hb_001821_090 |
0.1001838187 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 12 |
Hb_000307_210 |
0.1029503916 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 13 |
Hb_011512_090 |
0.1032614874 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 14 |
Hb_003177_060 |
0.103338808 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001123_050 |
0.1033849582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028487_010 |
0.103486274 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002477_090 |
0.1040126997 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000012_260 |
0.1042095875 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 19 |
Hb_000640_130 |
0.1047559592 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_002110_030 |
0.1048861787 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |