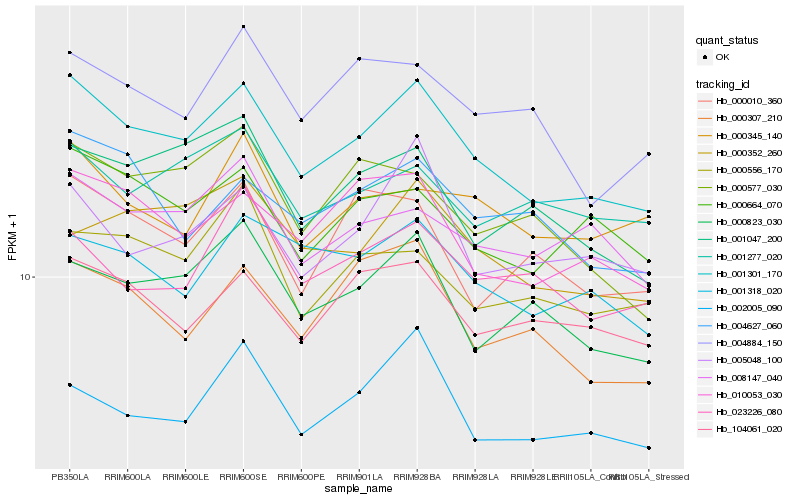
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_170 |
0.0 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 2 |
Hb_010053_030 |
0.079466841 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 3 |
Hb_000664_070 |
0.081950324 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 4 |
Hb_104061_020 |
0.0845330862 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 5 |
Hb_000823_030 |
0.0854744394 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 6 |
Hb_008147_040 |
0.0867849201 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 7 |
Hb_001318_020 |
0.087900741 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_004627_060 |
0.0906839374 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 9 |
Hb_005048_100 |
0.090821146 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 10 |
Hb_000307_210 |
0.091653542 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_001047_200 |
0.0920516121 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 12 |
Hb_004884_150 |
0.0926943824 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 13 |
Hb_001277_020 |
0.0936052961 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 14 |
Hb_000352_260 |
0.0944261058 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 15 |
Hb_000556_170 |
0.0962185278 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 16 |
Hb_023226_080 |
0.0965304646 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000345_140 |
0.0968506684 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 18 |
Hb_000010_360 |
0.097048359 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 19 |
Hb_000577_030 |
0.097481441 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 20 |
Hb_002005_090 |
0.0975209741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |