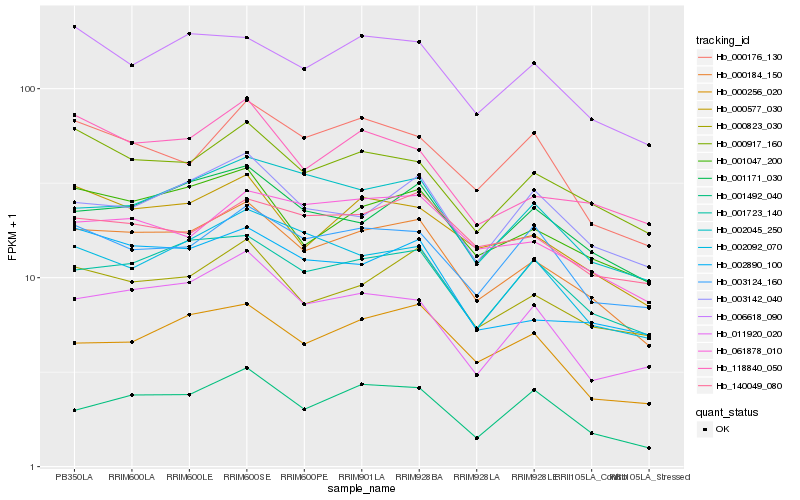
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_150 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 2 |
Hb_001047_200 |
0.0652617673 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 3 |
Hb_000577_030 |
0.0680319331 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 4 |
Hb_000823_030 |
0.0720776131 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 5 |
Hb_001723_140 |
0.0745189309 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_000917_160 |
0.0773787072 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006618_090 |
0.07797362 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 8 |
Hb_001171_030 |
0.0790458926 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 9 |
Hb_002045_250 |
0.0842067469 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 10 |
Hb_118840_050 |
0.0848895105 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 11 |
Hb_000256_020 |
0.0851704876 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 12 |
Hb_011920_020 |
0.0852239949 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 13 |
Hb_002890_100 |
0.0866863071 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002092_070 |
0.0887936591 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 15 |
Hb_003124_160 |
0.0891876081 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_061878_010 |
0.0893180292 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 17 |
Hb_000176_130 |
0.0909738524 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 18 |
Hb_001492_040 |
0.0911296476 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003142_040 |
0.0918561875 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 20 |
Hb_140049_080 |
0.0935094484 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |