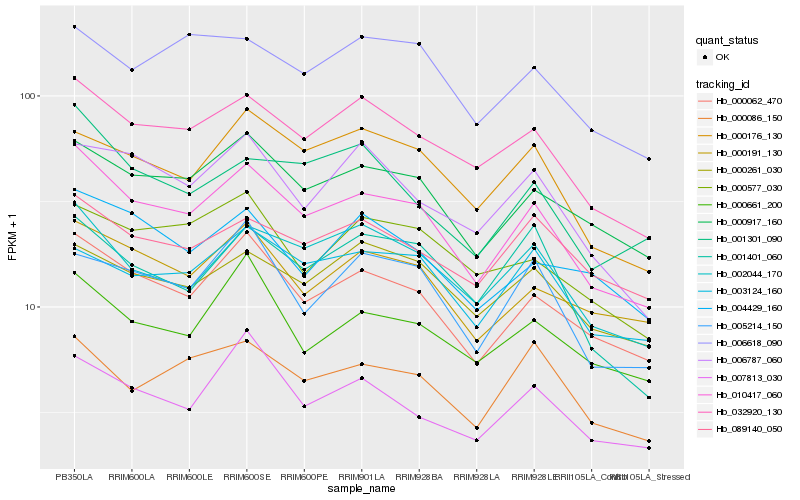
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010417_060 |
0.0 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 2 |
Hb_000062_470 |
0.0528556102 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 3 |
Hb_032920_130 |
0.0619038231 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 4 |
Hb_000917_160 |
0.0672442453 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002044_170 |
0.0734127997 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 6 |
Hb_000191_130 |
0.0780427205 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 7 |
Hb_000176_130 |
0.0853544538 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 8 |
Hb_000086_150 |
0.0869961398 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000661_200 |
0.0890158288 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001301_090 |
0.0905736363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000261_030 |
0.0912698533 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 12 |
Hb_003124_160 |
0.0914733875 |
- |
- |
dynamin, putative [Ricinus communis] |
| 13 |
Hb_007813_030 |
0.0918647741 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 14 |
Hb_004429_160 |
0.0919739557 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005214_150 |
0.0920397762 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 16 |
Hb_006787_060 |
0.0922334013 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_089140_050 |
0.0933892055 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 18 |
Hb_001401_060 |
0.0937682774 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 19 |
Hb_006618_090 |
0.0938049162 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 20 |
Hb_000577_030 |
0.0939430555 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |