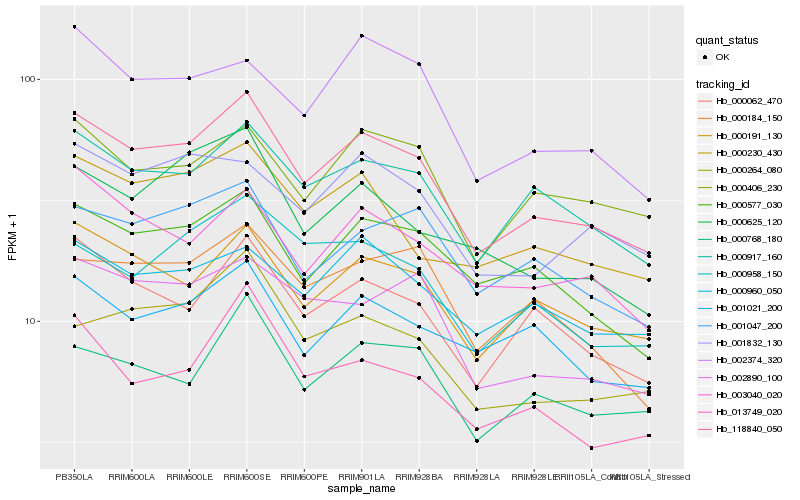
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118840_050 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 2 |
Hb_000917_160 |
0.0639660244 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000191_130 |
0.0656547223 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 4 |
Hb_000062_470 |
0.0703634311 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 5 |
Hb_000577_030 |
0.0714855978 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 6 |
Hb_000230_430 |
0.0779573937 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_001047_200 |
0.0795956655 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 8 |
Hb_003040_020 |
0.0803039969 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 9 |
Hb_002890_100 |
0.0808468925 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000768_180 |
0.0809097285 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 11 |
Hb_001021_200 |
0.0821077117 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 12 |
Hb_000406_230 |
0.0829137349 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 13 |
Hb_013749_020 |
0.0829316916 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000958_150 |
0.0833602417 |
- |
- |
Actin, putative [Ricinus communis] |
| 15 |
Hb_000960_050 |
0.0839344516 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 16 |
Hb_000264_080 |
0.0841999138 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 17 |
Hb_002374_320 |
0.0847160013 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 18 |
Hb_000184_150 |
0.0848895105 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 19 |
Hb_001832_130 |
0.0851071051 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 20 |
Hb_000625_120 |
0.0852334462 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |