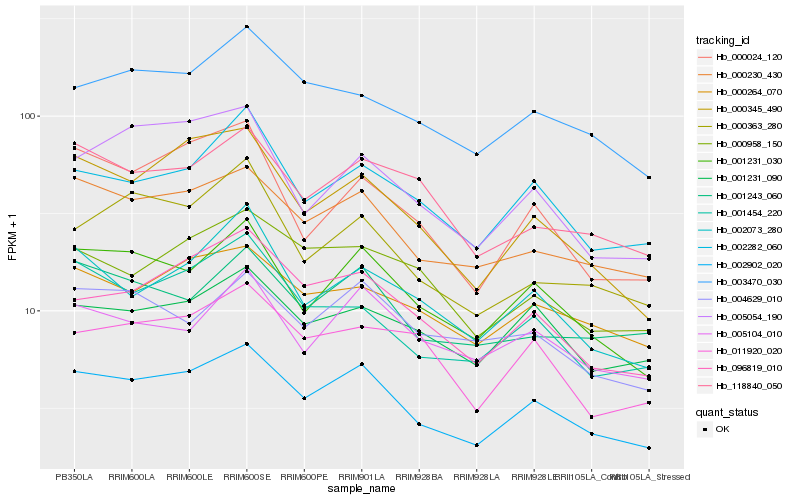
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002902_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 2 |
Hb_000230_430 |
0.0639657793 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_001231_030 |
0.0651135796 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 4 |
Hb_096819_010 |
0.08163573 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000345_490 |
0.0821647812 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 6 |
Hb_002282_060 |
0.0837761351 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000264_070 |
0.0872369687 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 8 |
Hb_001454_220 |
0.0908346897 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 9 |
Hb_003470_030 |
0.0916015971 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 10 |
Hb_001231_090 |
0.0917691948 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 11 |
Hb_118840_050 |
0.0923331123 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 12 |
Hb_002073_280 |
0.0938514856 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001243_060 |
0.0939468453 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 14 |
Hb_005104_010 |
0.0943579891 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 15 |
Hb_000958_150 |
0.0948652764 |
- |
- |
Actin, putative [Ricinus communis] |
| 16 |
Hb_000024_120 |
0.0951449736 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 17 |
Hb_004629_010 |
0.0964975865 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 18 |
Hb_011920_020 |
0.0972432463 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 19 |
Hb_005054_190 |
0.0981745238 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000363_280 |
0.0983859975 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |