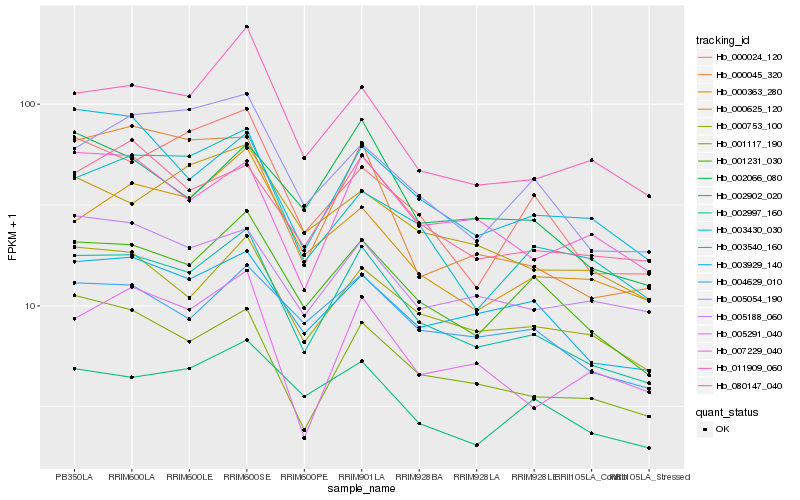
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002997_160 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001231_030 |
0.0774064657 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 3 |
Hb_000753_100 |
0.0785023025 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001117_190 |
0.0873113372 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 5 |
Hb_011909_060 |
0.0934262643 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 6 |
Hb_080147_040 |
0.0951243438 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 7 |
Hb_003929_140 |
0.0998083077 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_003430_030 |
0.1014529815 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 9 |
Hb_005054_190 |
0.1032815656 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002902_020 |
0.1039420833 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 11 |
Hb_004629_010 |
0.1051929272 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 12 |
Hb_000363_280 |
0.1055514562 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 13 |
Hb_007229_040 |
0.1056249674 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 14 |
Hb_002066_080 |
0.1062633098 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 15 |
Hb_000045_320 |
0.106796597 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003540_160 |
0.107045932 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 17 |
Hb_005291_040 |
0.1076671883 |
- |
- |
PREDICTED: gamma-gliadin [Jatropha curcas] |
| 18 |
Hb_000024_120 |
0.1088564451 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 19 |
Hb_005188_060 |
0.109060666 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 20 |
Hb_000625_120 |
0.1093542753 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |