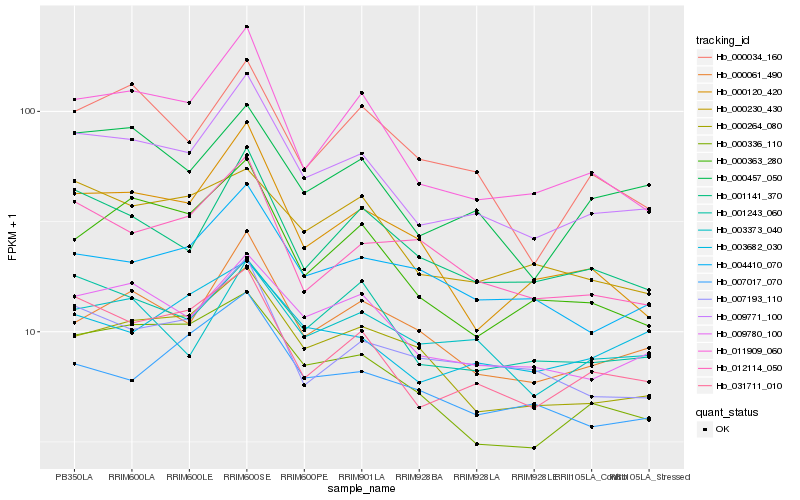
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 2 |
Hb_031711_010 |
0.065114114 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_009780_100 |
0.0710368559 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 4 |
Hb_011909_060 |
0.0729733311 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 5 |
Hb_001141_370 |
0.0773746149 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 6 |
Hb_000457_050 |
0.0817770613 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 7 |
Hb_000061_490 |
0.0820696925 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 8 |
Hb_001243_060 |
0.083162254 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 9 |
Hb_007193_110 |
0.0894186173 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 10 |
Hb_000264_080 |
0.0908573717 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 11 |
Hb_000363_280 |
0.0920725141 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 12 |
Hb_000336_110 |
0.0920903451 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000230_430 |
0.0935374078 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_004410_070 |
0.0938287279 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 15 |
Hb_000120_420 |
0.0948478745 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 16 |
Hb_003373_040 |
0.0975602915 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_000034_160 |
0.0981532532 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 18 |
Hb_012114_050 |
0.0982621341 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007017_070 |
0.0992993679 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003682_030 |
0.1007851121 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |