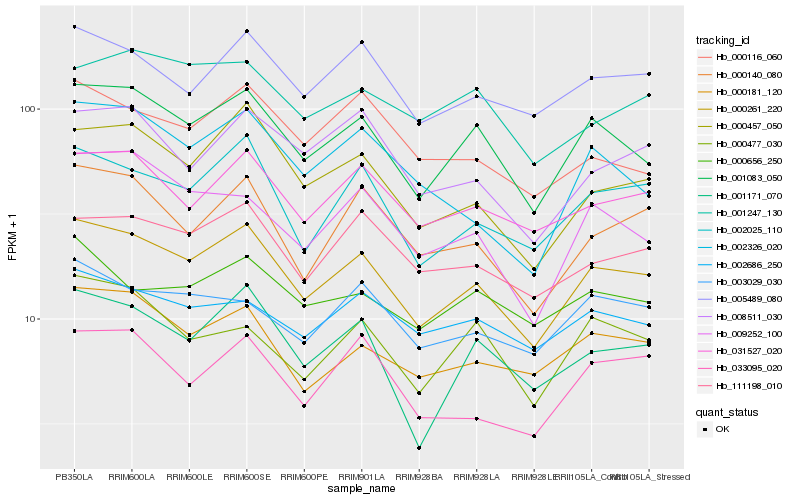
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_220 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 2 |
Hb_001083_050 |
0.0506460438 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 3 |
Hb_002025_110 |
0.062914894 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000457_050 |
0.0661048711 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 5 |
Hb_005489_080 |
0.0707323556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_031527_020 |
0.0732056153 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000140_080 |
0.0741445917 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000181_120 |
0.0753974729 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 9 |
Hb_111198_010 |
0.0763721579 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001171_070 |
0.0787421999 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_008511_030 |
0.0839816197 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 12 |
Hb_003029_030 |
0.0844730173 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002686_250 |
0.0857509706 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_009252_100 |
0.0860359777 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 15 |
Hb_033095_020 |
0.0868632663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000656_250 |
0.0882530652 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 17 |
Hb_000116_060 |
0.0884441143 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 18 |
Hb_000477_030 |
0.0899387912 |
- |
- |
PREDICTED: boron transporter 4-like [Jatropha curcas] |
| 19 |
Hb_002326_020 |
0.0916716396 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 20 |
Hb_001247_130 |
0.0919084024 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |