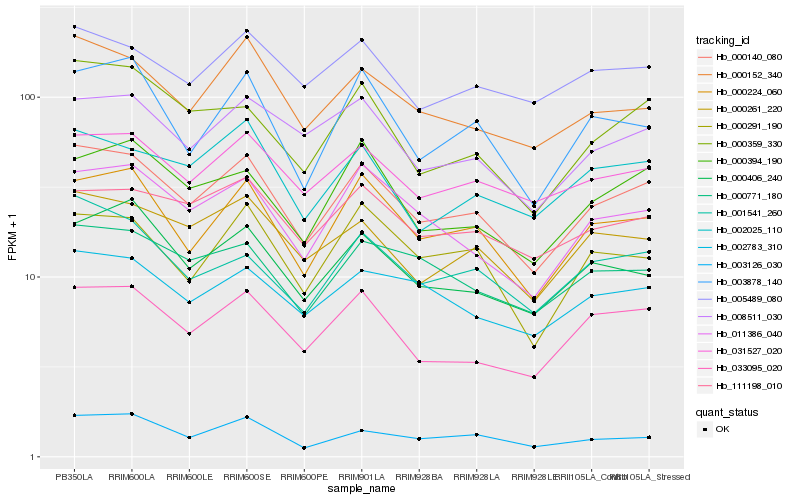
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_080 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000224_060 |
0.0632589004 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 3 |
Hb_033095_020 |
0.0679871426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011386_040 |
0.0730805246 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000261_220 |
0.0741445917 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 6 |
Hb_031527_020 |
0.075486953 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000152_340 |
0.0759414598 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 8 |
Hb_008511_030 |
0.0773447518 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 9 |
Hb_002025_110 |
0.0803917196 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000359_330 |
0.0819233777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000394_190 |
0.0820791273 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000406_240 |
0.082497838 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 13 |
Hb_002783_310 |
0.0832397732 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 14 |
Hb_001541_260 |
0.0838260312 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 15 |
Hb_000771_180 |
0.084431807 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 16 |
Hb_111198_010 |
0.0847083854 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000291_190 |
0.0848889899 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 18 |
Hb_005489_080 |
0.0852907821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003878_140 |
0.0861671329 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 20 |
Hb_003126_030 |
0.0862752769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |