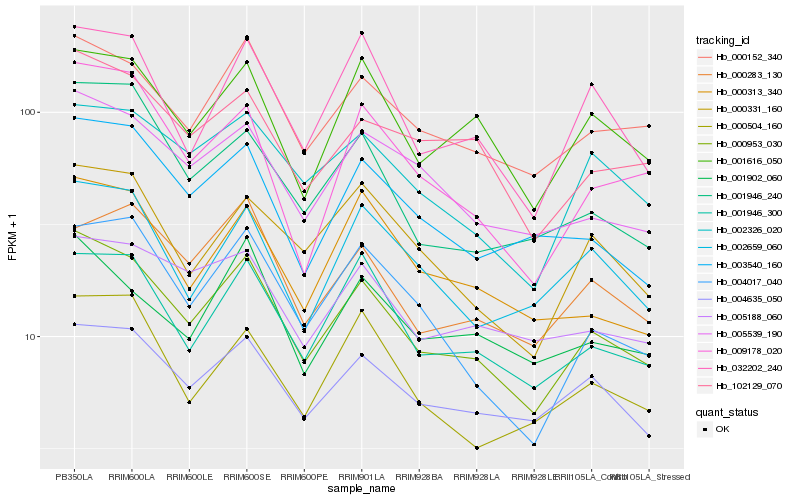
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000953_030 |
0.0 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000152_340 |
0.0742926018 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 3 |
Hb_001946_300 |
0.078653503 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 4 |
Hb_005539_190 |
0.0811215787 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 5 |
Hb_004635_050 |
0.0824156836 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 6 |
Hb_102129_070 |
0.0847117661 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 7 |
Hb_002326_020 |
0.0862157253 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 8 |
Hb_001616_050 |
0.0890082513 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 9 |
Hb_000331_160 |
0.0891674985 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 10 |
Hb_003540_160 |
0.0898075124 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 11 |
Hb_032202_240 |
0.0899943553 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 12 |
Hb_009178_020 |
0.0903968988 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 13 |
Hb_001946_240 |
0.0904160199 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 14 |
Hb_001902_060 |
0.0917361759 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 15 |
Hb_004017_040 |
0.0918081975 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 16 |
Hb_005188_060 |
0.0944283719 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 17 |
Hb_000313_340 |
0.0944570471 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 18 |
Hb_000283_130 |
0.0946995372 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 19 |
Hb_002659_060 |
0.0953126673 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 20 |
Hb_000504_160 |
0.0960515866 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |