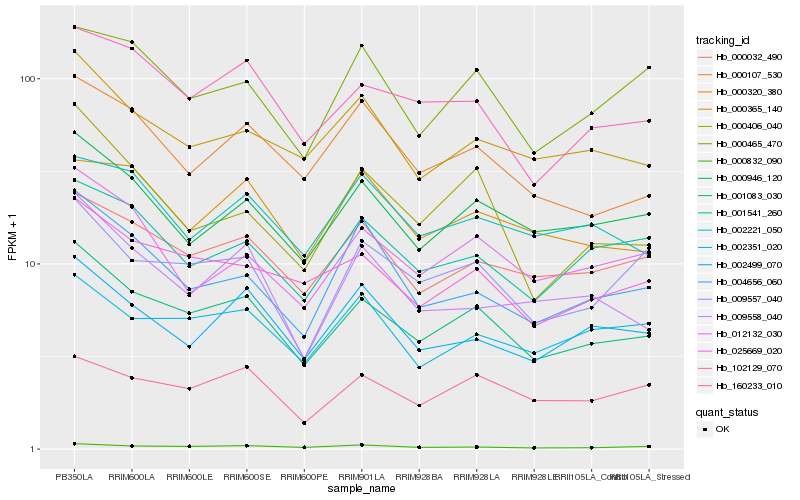
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001083_030 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 2 |
Hb_025669_020 |
0.0794644921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_102129_070 |
0.0834655913 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 4 |
Hb_000946_120 |
0.0862204687 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000032_490 |
0.0879059613 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 6 |
Hb_000365_140 |
0.0913846053 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 7 |
Hb_000107_530 |
0.0999105072 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_009557_040 |
0.1001411259 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 9 |
Hb_000832_090 |
0.1003771705 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 10 |
Hb_012132_030 |
0.1009110917 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 11 |
Hb_001541_260 |
0.1022977404 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 12 |
Hb_002499_070 |
0.1045153924 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 13 |
Hb_160233_010 |
0.1062344009 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 14 |
Hb_002351_020 |
0.1093395159 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 15 |
Hb_009558_040 |
0.1097044772 |
transcription factor |
TF Family: MYB |
hypothetical protein PHAVU_008G102000g [Phaseolus vulgaris] |
| 16 |
Hb_004656_060 |
0.1101836282 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 17 |
Hb_000406_040 |
0.1105340997 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 18 |
Hb_002221_050 |
0.1115044865 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000465_470 |
0.1115742297 |
- |
- |
unknown [Picea sitchensis] |
| 20 |
Hb_000320_380 |
0.113370547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |