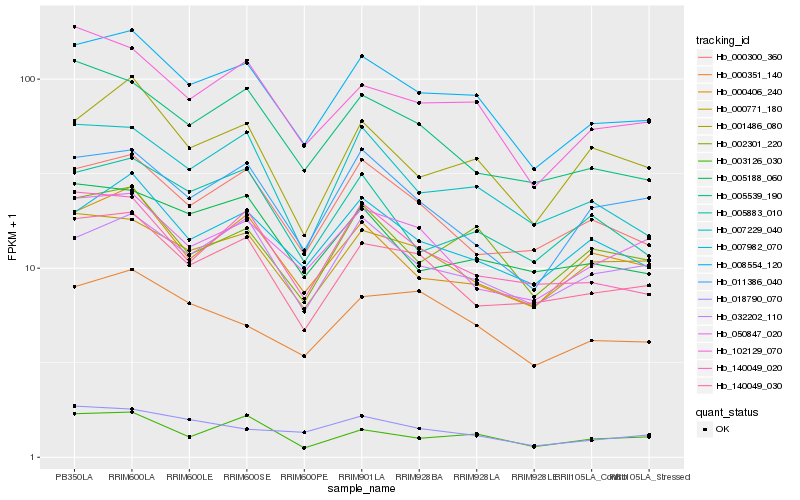
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008554_120 |
0.0 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 2 |
Hb_002301_220 |
0.0720951387 |
transcription factor |
TF Family: C3H |
PREDICTED: protein FRIGIDA-ESSENTIAL 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_102129_070 |
0.0750216341 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 4 |
Hb_007982_070 |
0.0756075946 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 5 |
Hb_140049_030 |
0.077203757 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 6 |
Hb_007229_040 |
0.0781241567 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 7 |
Hb_000771_180 |
0.0801811856 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_140049_020 |
0.0812471383 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 9 |
Hb_032202_110 |
0.0817164198 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 10 |
Hb_000406_240 |
0.0827957725 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 11 |
Hb_000300_360 |
0.0837191747 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 12 |
Hb_001486_080 |
0.083903096 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 13 |
Hb_005188_060 |
0.0878640959 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_005883_010 |
0.0883838797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003126_030 |
0.0886827824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005539_190 |
0.089084936 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 17 |
Hb_050847_020 |
0.0894632824 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_011386_040 |
0.0895188634 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000351_140 |
0.0895688541 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 9A-like isoform X3 [Pyrus x bretschneideri] |
| 20 |
Hb_018790_070 |
0.0919570973 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |