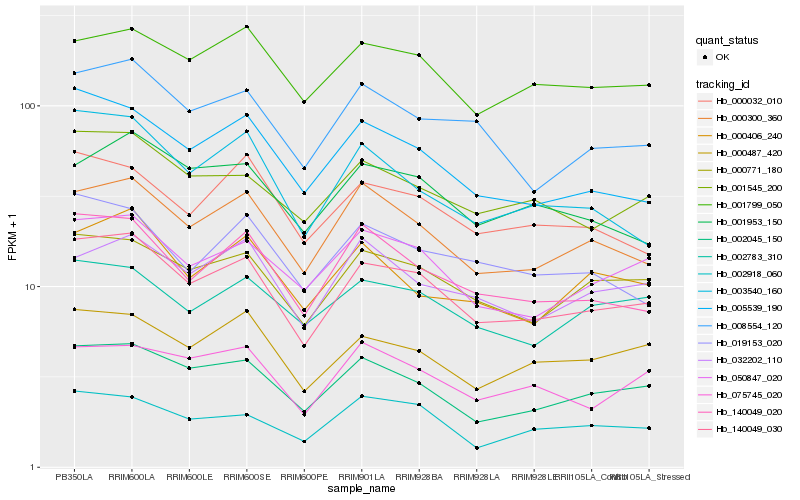
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140049_030 |
0.0 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 2 |
Hb_002045_150 |
0.0637727504 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 3 |
Hb_000771_180 |
0.0651733729 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 4 |
Hb_000300_360 |
0.0687810368 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 5 |
Hb_050847_020 |
0.0710432847 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_140049_020 |
0.0740591784 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 7 |
Hb_001545_200 |
0.0741410951 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 8 |
Hb_005539_190 |
0.0741554263 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 9 |
Hb_000487_420 |
0.0759815249 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 10 |
Hb_008554_120 |
0.077203757 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 11 |
Hb_001799_050 |
0.0815360482 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 12 |
Hb_000032_010 |
0.0856351383 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 13 |
Hb_002918_060 |
0.0859249204 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 14 |
Hb_002783_310 |
0.086953638 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 15 |
Hb_001953_150 |
0.0873358964 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 16 |
Hb_075745_020 |
0.088867827 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 17 |
Hb_032202_110 |
0.08956399 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_000406_240 |
0.0905346575 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_019153_020 |
0.0916163863 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003540_160 |
0.0916180685 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |