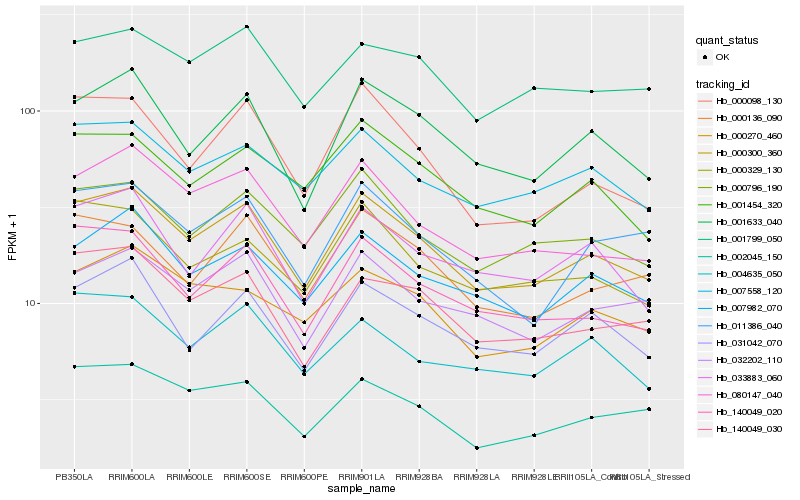
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_360 |
0.0 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 2 |
Hb_007982_070 |
0.0615266096 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 3 |
Hb_001633_040 |
0.0622561901 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 4 |
Hb_000796_190 |
0.0629577133 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 5 |
Hb_140049_020 |
0.0633818151 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 6 |
Hb_080147_040 |
0.0645797902 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 7 |
Hb_007558_120 |
0.0672135952 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 8 |
Hb_032202_110 |
0.0685087098 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 9 |
Hb_140049_030 |
0.0687810368 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 10 |
Hb_031042_070 |
0.0709568965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002045_150 |
0.0711469805 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 12 |
Hb_001454_320 |
0.0714208899 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000329_130 |
0.0720923173 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 14 |
Hb_033883_060 |
0.072374933 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 15 |
Hb_000136_090 |
0.0731921906 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 16 |
Hb_000270_460 |
0.0744489797 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 17 |
Hb_000098_130 |
0.0746512113 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 18 |
Hb_001799_050 |
0.0758263759 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 19 |
Hb_011386_040 |
0.0760949243 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004635_050 |
0.0787531596 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |