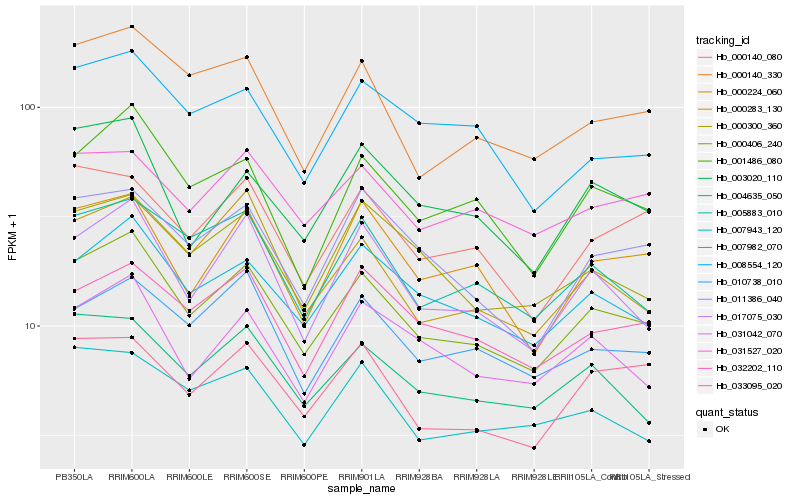
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000406_240 |
0.0 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 2 |
Hb_007982_070 |
0.062191511 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 3 |
Hb_017075_030 |
0.0689832488 |
- |
- |
helicase, putative [Ricinus communis] |
| 4 |
Hb_001486_080 |
0.069011351 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 5 |
Hb_005883_010 |
0.0736795853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000140_330 |
0.0738574557 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 7 |
Hb_004635_050 |
0.0747000441 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 8 |
Hb_000300_360 |
0.0788500399 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 9 |
Hb_031042_070 |
0.0792617979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010738_010 |
0.0792672474 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_000283_130 |
0.0795433618 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 12 |
Hb_032202_110 |
0.0795478573 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 13 |
Hb_003020_110 |
0.0803516054 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 14 |
Hb_011386_040 |
0.0807235085 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000224_060 |
0.081079087 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 16 |
Hb_000140_080 |
0.082497838 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_008554_120 |
0.0827957725 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 18 |
Hb_007943_120 |
0.0852019192 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 19 |
Hb_033095_020 |
0.0854687155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_031527_020 |
0.0861516355 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |