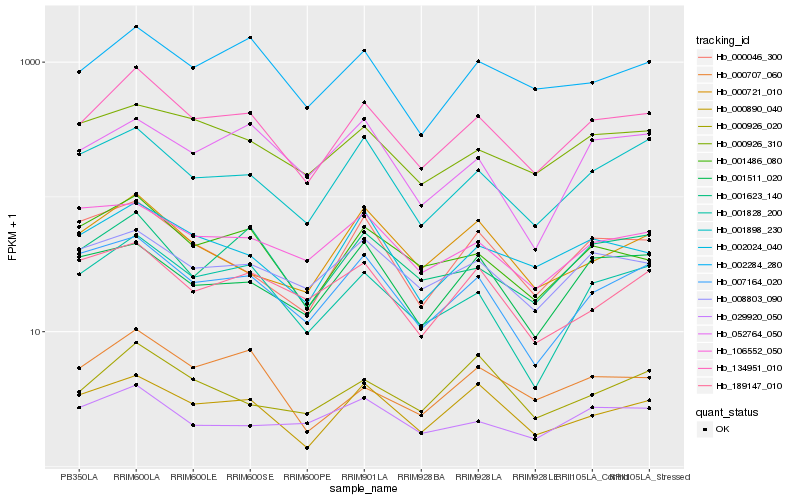
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134951_010 |
0.0 |
- |
- |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 2 |
Hb_001486_080 |
0.0896745793 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 3 |
Hb_001828_200 |
0.0920632582 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 4 |
Hb_001898_230 |
0.0977207658 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002024_040 |
0.0987113502 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 6 |
Hb_002284_280 |
0.0997233062 |
transcription factor |
TF Family: HMG |
high mobility group protein [Hevea brasiliensis] |
| 7 |
Hb_001623_140 |
0.1018968371 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 8 |
Hb_000890_040 |
0.1035072764 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g15720 [Jatropha curcas] |
| 9 |
Hb_007164_020 |
0.1036160956 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 10 |
Hb_008803_090 |
0.1046659901 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 11 |
Hb_000926_310 |
0.1053523117 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 12 |
Hb_000046_300 |
0.1112088383 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 13 |
Hb_000707_060 |
0.1118157303 |
- |
- |
- |
| 14 |
Hb_000721_010 |
0.1175272296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001511_020 |
0.121035426 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |
| 16 |
Hb_106552_050 |
0.1211582215 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 17 |
Hb_189147_010 |
0.1227791424 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_029920_050 |
0.1230165348 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 19 |
Hb_052764_050 |
0.123284053 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 20 |
Hb_000926_020 |
0.1241908173 |
- |
- |
- |