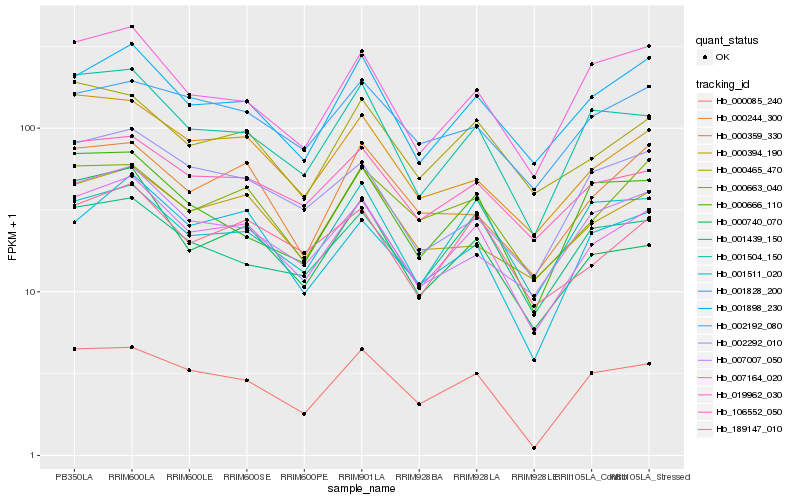
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007164_020 |
0.0 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 2 |
Hb_189147_010 |
0.0751649467 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001504_150 |
0.0753125488 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 4 |
Hb_001898_230 |
0.0761003311 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 5 |
Hb_106552_050 |
0.0794147673 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 6 |
Hb_000085_240 |
0.0842044224 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 7 |
Hb_000359_330 |
0.0846729576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001828_200 |
0.0863123414 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 9 |
Hb_000465_470 |
0.0865706127 |
- |
- |
unknown [Picea sitchensis] |
| 10 |
Hb_019962_030 |
0.0882271438 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 11 |
Hb_000663_040 |
0.0906275171 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000740_070 |
0.0912345413 |
- |
- |
- |
| 13 |
Hb_007007_050 |
0.0915283527 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 14 |
Hb_000394_190 |
0.0919979072 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_001439_150 |
0.0965877713 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000666_110 |
0.098861811 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 17 |
Hb_002292_010 |
0.0991069285 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 18 |
Hb_002192_080 |
0.0991746604 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 19 |
Hb_000244_300 |
0.0993728798 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001511_020 |
0.0999171011 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |