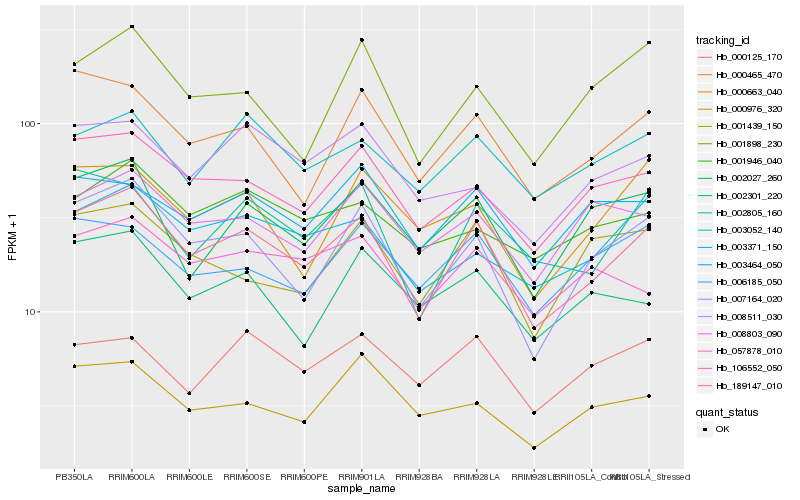
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189147_010 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007164_020 |
0.0751649467 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 3 |
Hb_106552_050 |
0.0869220086 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 4 |
Hb_002805_160 |
0.0955635898 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 5 |
Hb_000465_470 |
0.0974537403 |
- |
- |
unknown [Picea sitchensis] |
| 6 |
Hb_003371_150 |
0.1017509358 |
- |
- |
PREDICTED: serine/threonine-protein kinase 10 [Jatropha curcas] |
| 7 |
Hb_003052_140 |
0.1031001977 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 8 |
Hb_002027_260 |
0.1041039133 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 9 |
Hb_003464_050 |
0.1092669349 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 10 |
Hb_001439_150 |
0.1101410695 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 11 |
Hb_006185_050 |
0.1107150458 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 12 |
Hb_001898_230 |
0.1107867186 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 13 |
Hb_057878_010 |
0.1110195593 |
- |
- |
hypothetical protein VITISV_036304 [Vitis vinifera] |
| 14 |
Hb_008803_090 |
0.1121273076 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 15 |
Hb_000125_170 |
0.112340995 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 16 |
Hb_001946_040 |
0.1133147243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008511_030 |
0.1138593223 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 18 |
Hb_000663_040 |
0.1144139358 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000976_320 |
0.1148766384 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 20 |
Hb_002301_220 |
0.1153651828 |
transcription factor |
TF Family: C3H |
PREDICTED: protein FRIGIDA-ESSENTIAL 1-like isoform X1 [Jatropha curcas] |