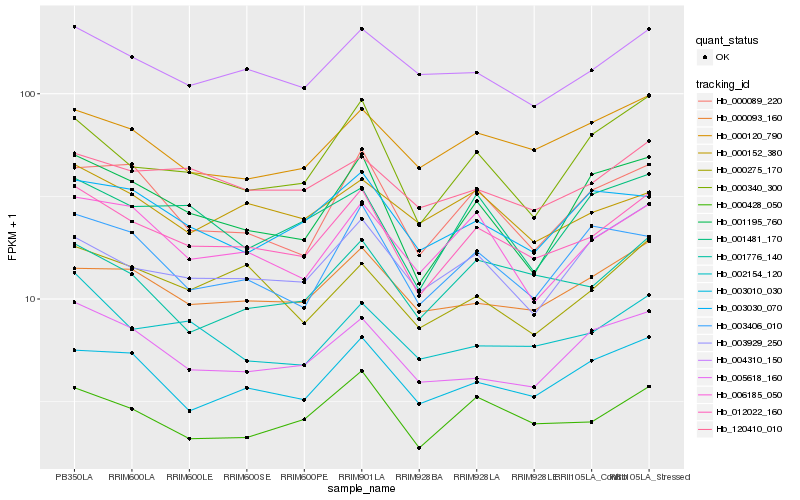
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012022_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 2 |
Hb_000428_050 |
0.070149393 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004310_150 |
0.0757599067 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 4 |
Hb_003010_030 |
0.0763649599 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 5 |
Hb_000093_160 |
0.0776417769 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_006185_050 |
0.0780446276 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 7 |
Hb_001195_760 |
0.0785552367 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_000089_220 |
0.0785756089 |
- |
- |
PREDICTED: protein FAM133-like [Populus euphratica] |
| 9 |
Hb_000120_790 |
0.0787473496 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 10 |
Hb_002154_120 |
0.0796664397 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 11 |
Hb_120410_010 |
0.080678808 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 12 |
Hb_001776_140 |
0.0807162469 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000340_300 |
0.0818887302 |
- |
- |
methyl-CpG-binding domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_001481_170 |
0.0835523035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005618_160 |
0.0837409673 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 16 |
Hb_003030_070 |
0.0842656581 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 17 |
Hb_000275_170 |
0.0845654615 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 18 |
Hb_000152_380 |
0.0860067903 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 19 |
Hb_003406_010 |
0.0869112513 |
- |
- |
putative ATP-dependent RNA helicase DHX36 [Morus notabilis] |
| 20 |
Hb_003929_250 |
0.0869318727 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |