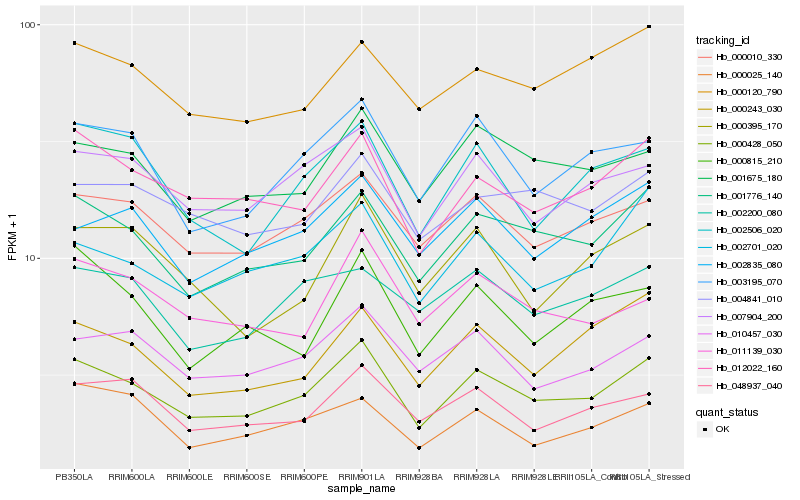
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000428_050 |
0.0 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001776_140 |
0.0565100471 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_012022_160 |
0.070149393 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 4 |
Hb_000010_330 |
0.072191485 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 5 |
Hb_007904_200 |
0.0751432603 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_003195_070 |
0.0755363777 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_001675_180 |
0.0768668117 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 8 |
Hb_002701_020 |
0.0798265329 |
- |
- |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 9 |
Hb_002506_020 |
0.0819589263 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_010457_030 |
0.0819748814 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 11 |
Hb_048937_040 |
0.0831161025 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_000243_030 |
0.086493693 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 13 |
Hb_000120_790 |
0.0873143245 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 14 |
Hb_004841_010 |
0.0874690367 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002835_080 |
0.0881897089 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_011139_030 |
0.0921562117 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 17 |
Hb_002200_080 |
0.0922538126 |
- |
- |
PREDICTED: uncharacterized protein C57A10.07 [Jatropha curcas] |
| 18 |
Hb_000395_170 |
0.0923522177 |
- |
- |
PREDICTED: probable transcriptional regulatory protein At2g25830 [Jatropha curcas] |
| 19 |
Hb_000025_140 |
0.0923672621 |
- |
- |
las1-like family protein [Populus trichocarpa] |
| 20 |
Hb_000815_210 |
0.0934097566 |
- |
- |
uncoupling protein 3, partial [Manihot esculenta] |