| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
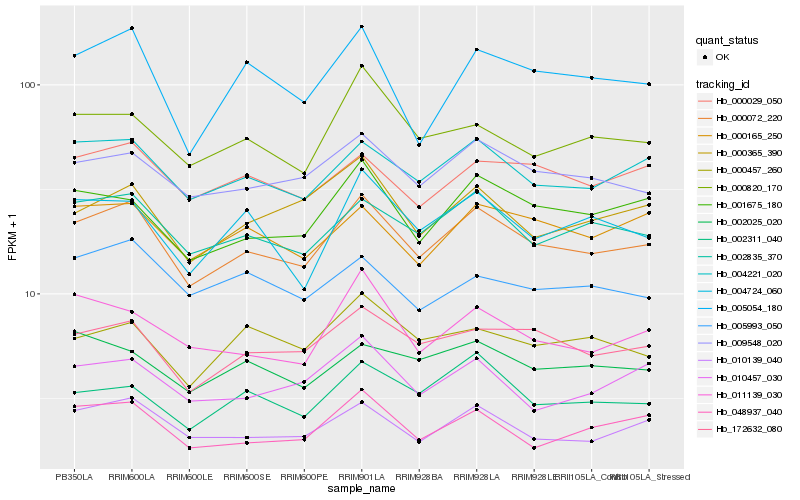
Hb_000072_220 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02490, mitochondrial [Jatropha curcas] |
| 2 |
Hb_004221_020 |
0.054001299 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_002835_370 |
0.0554390818 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 4 |
Hb_010139_040 |
0.0574741995 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_172632_080 |
0.0600530621 |
- |
- |
PREDICTED: uncharacterized protein LOC105141138 isoform X1 [Populus euphratica] |
| 6 |
Hb_009548_020 |
0.0615606215 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 7 |
Hb_001675_180 |
0.064226718 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 8 |
Hb_000165_250 |
0.0704254194 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 9 |
Hb_048937_040 |
0.0708941807 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 10 |
Hb_000029_050 |
0.0718810791 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_002311_040 |
0.0760929787 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 12 |
Hb_005054_180 |
0.0765265573 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 13 |
Hb_004724_060 |
0.076958859 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000365_390 |
0.0771981055 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 15 |
Hb_000820_170 |
0.0783446386 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 16 |
Hb_011139_030 |
0.0796289089 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 17 |
Hb_002025_020 |
0.0799298149 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 18 |
Hb_010457_030 |
0.0805829448 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 19 |
Hb_000457_260 |
0.0806665288 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005993_050 |
0.0809228727 |
- |
- |
Protein AFR, putative [Ricinus communis] |