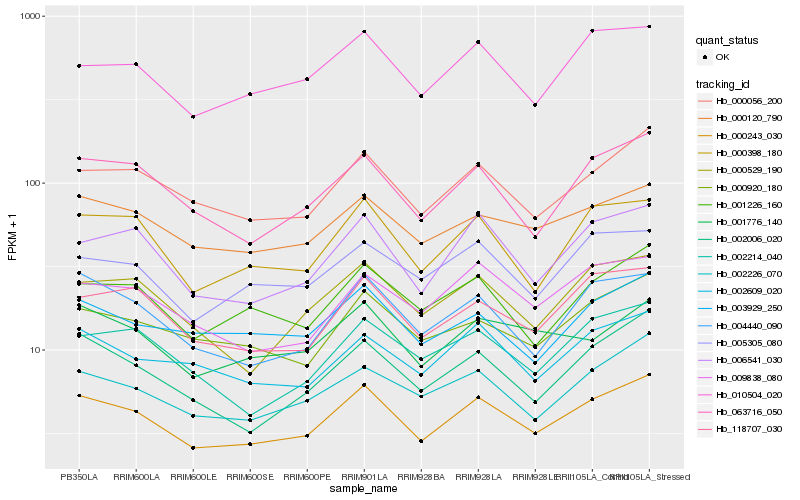
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_030 |
0.0 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 2 |
Hb_000056_200 |
0.0583921643 |
- |
- |
40S ribosomal S9-2 -like protein [Gossypium arboreum] |
| 3 |
Hb_063716_050 |
0.0632231667 |
- |
- |
PREDICTED: uncharacterized protein LOC105634866 [Jatropha curcas] |
| 4 |
Hb_006541_030 |
0.0644386603 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 5 |
Hb_000398_180 |
0.0652145407 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 6 |
Hb_002226_070 |
0.0659598095 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004440_090 |
0.0660064579 |
- |
- |
PREDICTED: phosphopantetheine adenylyltransferase [Jatropha curcas] |
| 8 |
Hb_000120_790 |
0.0683011905 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 9 |
Hb_118707_030 |
0.0687388873 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 19a-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_002006_020 |
0.0703921834 |
- |
- |
PREDICTED: uncharacterized protein LOC105650638 [Jatropha curcas] |
| 11 |
Hb_010504_020 |
0.0703923007 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 12 |
Hb_001226_160 |
0.0713356185 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000920_180 |
0.0726157861 |
- |
- |
PREDICTED: F-box protein SKIP5 [Jatropha curcas] |
| 14 |
Hb_002609_020 |
0.0738170288 |
- |
- |
PREDICTED: phosphatidylinositol N-acetylglucosaminyltransferase subunit P-like [Jatropha curcas] |
| 15 |
Hb_000529_190 |
0.0743562074 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1-like [Jatropha curcas] |
| 16 |
Hb_009838_080 |
0.0769758171 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 17 |
Hb_005305_080 |
0.0769763366 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003929_250 |
0.0775336573 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 19 |
Hb_002214_040 |
0.078295559 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_001776_140 |
0.0795587901 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |