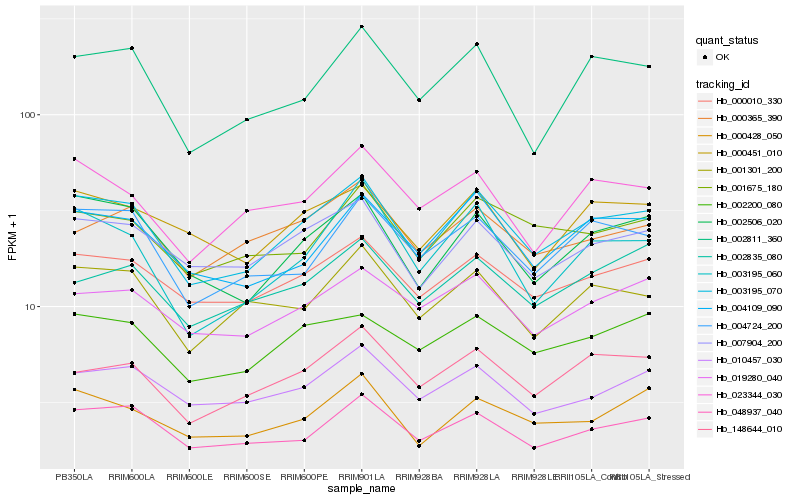
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003195_070 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002506_020 |
0.0592182025 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000010_330 |
0.0609672969 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 4 |
Hb_007904_200 |
0.063423814 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_048937_040 |
0.0650139452 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_004109_090 |
0.0669635449 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_002200_080 |
0.0674024856 |
- |
- |
PREDICTED: uncharacterized protein C57A10.07 [Jatropha curcas] |
| 8 |
Hb_148644_010 |
0.0692148153 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004724_200 |
0.0720175022 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 10 |
Hb_002811_360 |
0.0725397197 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 11 |
Hb_001301_200 |
0.0727555653 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 12 |
Hb_019280_040 |
0.0730689198 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_010457_030 |
0.0733900147 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_000451_010 |
0.0745048428 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 15 |
Hb_000428_050 |
0.0755363777 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003195_060 |
0.077156292 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 17 |
Hb_001675_180 |
0.0778931472 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 18 |
Hb_023344_030 |
0.0781358412 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 19 |
Hb_002835_080 |
0.0813252006 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000365_390 |
0.0820890292 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |