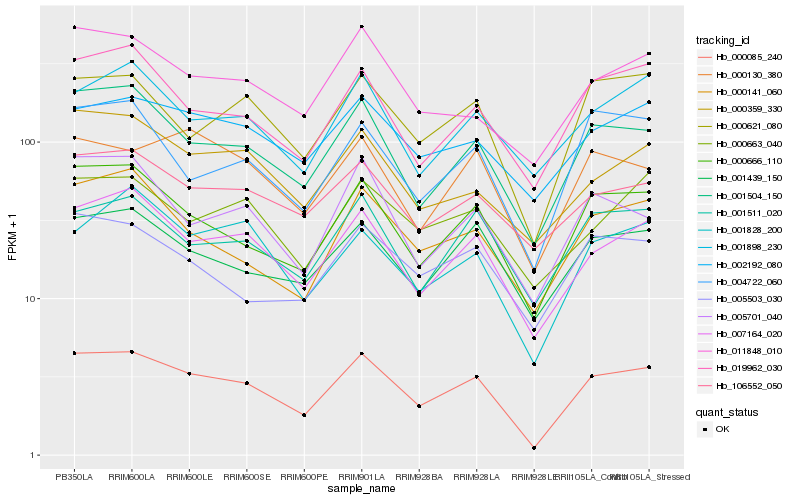
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_240 |
0.0 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 2 |
Hb_000666_110 |
0.0840931319 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 3 |
Hb_007164_020 |
0.0842044224 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 4 |
Hb_001504_150 |
0.0845912724 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 5 |
Hb_000621_080 |
0.1018645115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_019962_030 |
0.1059927303 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 7 |
Hb_001439_150 |
0.1067188567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000141_060 |
0.1085954861 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 9 |
Hb_001511_020 |
0.1107207994 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |
| 10 |
Hb_002192_080 |
0.1116400059 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 11 |
Hb_004722_060 |
0.1145441576 |
- |
- |
PREDICTED: protein CLP1 homolog [Jatropha curcas] |
| 12 |
Hb_005701_040 |
0.1148443085 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_106552_050 |
0.1148736827 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 14 |
Hb_001828_200 |
0.1158998637 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 15 |
Hb_011848_010 |
0.1160670923 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 16 |
Hb_005503_030 |
0.1172003216 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000130_380 |
0.1173514636 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000359_330 |
0.1173826563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000663_040 |
0.1186064775 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001898_230 |
0.1207917773 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |