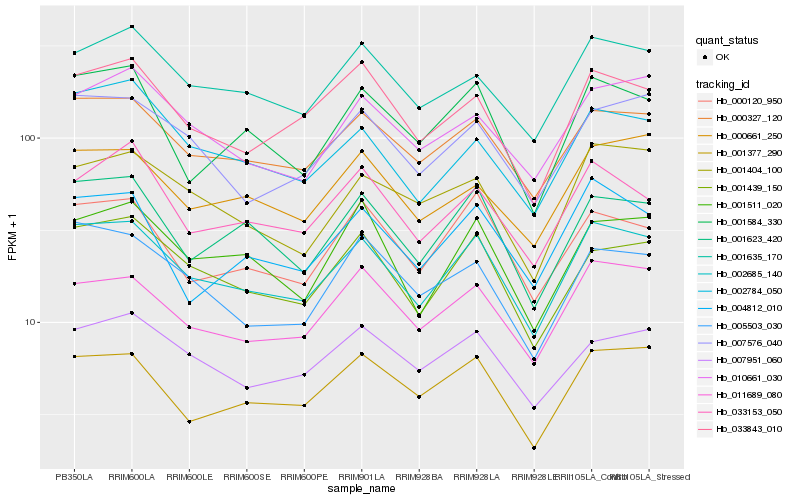
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002685_140 |
0.0 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 2 |
Hb_000327_120 |
0.0478061185 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 3 |
Hb_001439_150 |
0.0601576472 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 4 |
Hb_011689_080 |
0.0641800218 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 5 |
Hb_001623_420 |
0.0666518618 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 6 |
Hb_001635_170 |
0.0668487304 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 7 |
Hb_007576_040 |
0.0706690054 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 8 |
Hb_000120_950 |
0.0724132188 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 9 |
Hb_002784_050 |
0.0741636619 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 [Jatropha curcas] |
| 10 |
Hb_001377_290 |
0.074204677 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase accessory protein, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001511_020 |
0.0742868169 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |
| 12 |
Hb_007951_060 |
0.0747448129 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_033843_010 |
0.0748349404 |
- |
- |
PREDICTED: cell division control protein 2 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_005503_030 |
0.0772025762 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001584_330 |
0.0776787017 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 16 |
Hb_001404_100 |
0.0776892423 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K [Gossypium raimondii] |
| 17 |
Hb_010661_030 |
0.078107043 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 18 |
Hb_004812_010 |
0.0782849654 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD6 [Jatropha curcas] |
| 19 |
Hb_000661_250 |
0.0785820261 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 20 |
Hb_033153_050 |
0.0786805127 |
transcription factor |
TF Family: VOZ |
conserved hypothetical protein [Ricinus communis] |