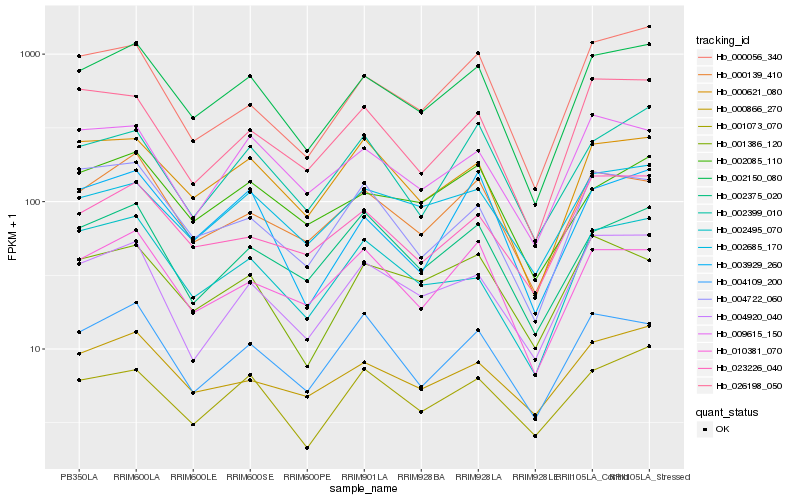
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_080 |
0.0 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 2 |
Hb_000139_410 |
0.0779029734 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 3 |
Hb_000621_080 |
0.077980632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002495_070 |
0.0858619928 |
- |
- |
PREDICTED: uncharacterized protein LOC105634731 [Jatropha curcas] |
| 5 |
Hb_010381_070 |
0.0860457932 |
- |
- |
PREDICTED: protein STU1-like [Jatropha curcas] |
| 6 |
Hb_002085_110 |
0.0876509528 |
- |
- |
Protein transport protein Sec61 subunit beta, putative [Ricinus communis] |
| 7 |
Hb_002685_170 |
0.0881600738 |
- |
- |
hypothetical protein EUTSA_v10019281mg [Eutrema salsugineum] |
| 8 |
Hb_000866_270 |
0.0894728533 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 9 |
Hb_002375_020 |
0.0897937268 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 10 |
Hb_000056_340 |
0.0900890814 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 11 |
Hb_001073_070 |
0.0902103292 |
- |
- |
PREDICTED: uncharacterized protein LOC105644546 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003929_260 |
0.0905373393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004109_200 |
0.0907085487 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_023226_040 |
0.0932548546 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_004920_040 |
0.0940043975 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 16 |
Hb_009615_150 |
0.0945731651 |
- |
- |
PREDICTED: transcription elongation factor SPT4 homolog 2 [Jatropha curcas] |
| 17 |
Hb_001386_120 |
0.0967731359 |
- |
- |
PREDICTED: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 18 |
Hb_004722_060 |
0.0975348873 |
- |
- |
PREDICTED: protein CLP1 homolog [Jatropha curcas] |
| 19 |
Hb_026198_050 |
0.0979287599 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 20 |
Hb_002399_010 |
0.0982378108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |