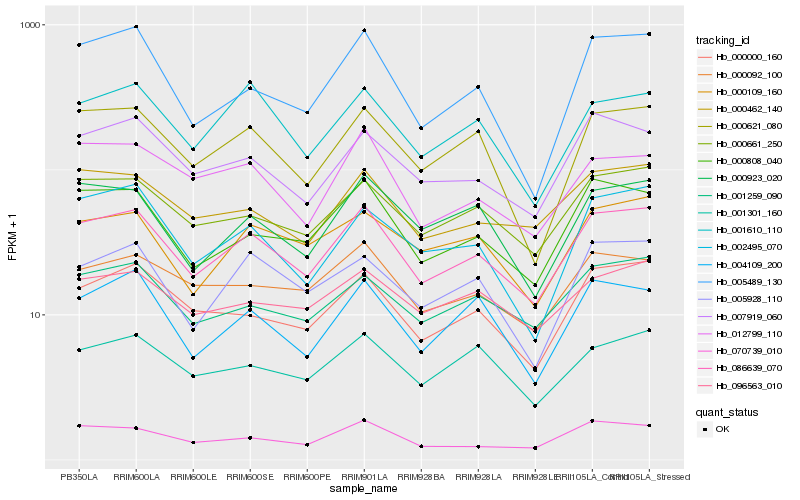
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086639_070 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 2 |
Hb_000661_250 |
0.0687191784 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 3 |
Hb_000808_040 |
0.0709540456 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 3 [Jatropha curcas] |
| 4 |
Hb_000000_160 |
0.0754262298 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 5 |
Hb_070739_010 |
0.0766004655 |
- |
- |
PREDICTED: probable aminotransferase ACS12 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001259_090 |
0.0767458806 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 7 |
Hb_007919_060 |
0.0769343079 |
- |
- |
PREDICTED: UPF0047 protein C4A8.02c [Jatropha curcas] |
| 8 |
Hb_000621_080 |
0.0781024388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000462_140 |
0.081098063 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 10 |
Hb_000923_020 |
0.0837621533 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 11 |
Hb_000092_100 |
0.0844891814 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 12 |
Hb_001610_110 |
0.0846129263 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 13 |
Hb_000109_160 |
0.084802503 |
- |
- |
hypothetical protein PRUPE_ppa012786mg [Prunus persica] |
| 14 |
Hb_001301_160 |
0.0856609536 |
- |
- |
PREDICTED: CRS2-associated factor 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_005928_110 |
0.0861100458 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 16 |
Hb_004109_200 |
0.0864482016 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_096563_010 |
0.0889380115 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 18 |
Hb_012799_110 |
0.0890147809 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 19 |
Hb_005489_130 |
0.0890689669 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 20 |
Hb_002495_070 |
0.0896989051 |
- |
- |
PREDICTED: uncharacterized protein LOC105634731 [Jatropha curcas] |