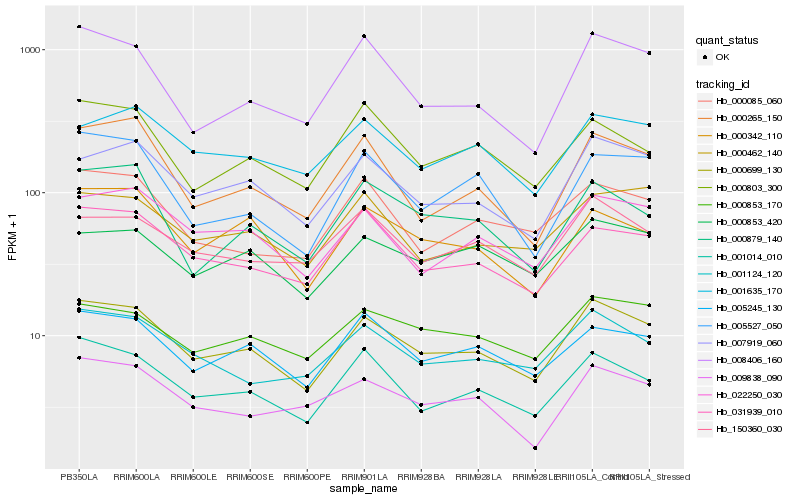
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000699_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007919_060 |
0.0692901905 |
- |
- |
PREDICTED: UPF0047 protein C4A8.02c [Jatropha curcas] |
| 3 |
Hb_001014_010 |
0.071951745 |
- |
- |
PREDICTED: uncharacterized protein LOC105647311 [Jatropha curcas] |
| 4 |
Hb_022250_030 |
0.0776642786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005245_130 |
0.0805985155 |
- |
- |
PREDICTED: uncharacterized protein LOC105643730 [Jatropha curcas] |
| 6 |
Hb_001124_120 |
0.0807515856 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 7 |
Hb_031939_010 |
0.0827505149 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000853_170 |
0.083761566 |
- |
- |
PREDICTED: programmed cell death protein 2 isoform X3 [Populus euphratica] |
| 9 |
Hb_008406_160 |
0.0879478889 |
- |
- |
PREDICTED: ADP,ATP carrier protein 3, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000342_110 |
0.0896316128 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 11 |
Hb_000265_150 |
0.0901154437 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 12 |
Hb_150360_030 |
0.0911528585 |
- |
- |
PREDICTED: F-box/WD-40 repeat-containing protein At3g52030-like [Jatropha curcas] |
| 13 |
Hb_000085_060 |
0.0916850661 |
- |
- |
PREDICTED: uncharacterized protein LOC105633020 [Jatropha curcas] |
| 14 |
Hb_000462_140 |
0.0917089822 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 15 |
Hb_001635_170 |
0.0918462066 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 16 |
Hb_000879_140 |
0.0934160203 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0283697-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_009838_090 |
0.0935773085 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000853_420 |
0.0945544523 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_005527_050 |
0.0946722359 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 20 |
Hb_000803_300 |
0.0948510305 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |