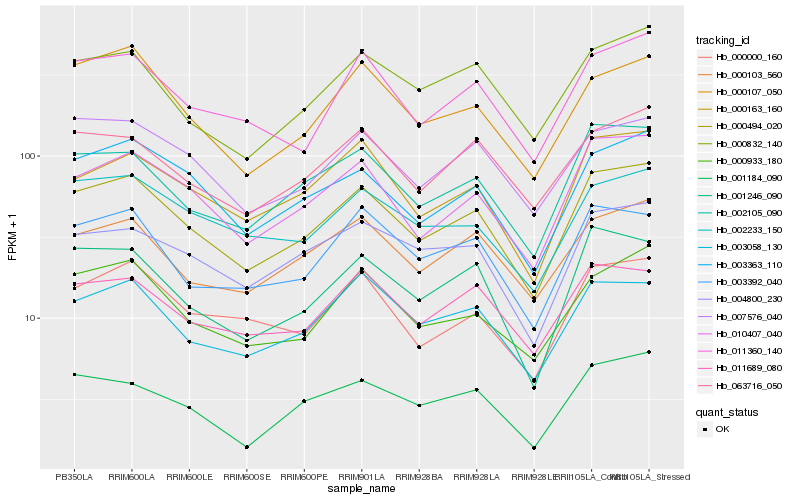
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000494_020 |
0.0 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 5A [Jatropha curcas] |
| 2 |
Hb_010407_040 |
0.0530873522 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_002105_090 |
0.0590303154 |
- |
- |
Rab4 [Hevea brasiliensis] |
| 4 |
Hb_003363_110 |
0.0635510273 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_000933_180 |
0.0650540299 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000163_160 |
0.0676116086 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 7 |
Hb_001246_090 |
0.0695975088 |
- |
- |
PREDICTED: acyl carrier protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_011360_140 |
0.0710017677 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 9 |
Hb_000103_560 |
0.0714042476 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 10 |
Hb_003058_130 |
0.0715535241 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A isoform X1 [Jatropha curcas] |
| 11 |
Hb_000000_160 |
0.0717277819 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 12 |
Hb_003392_040 |
0.0720912172 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Jatropha curcas] |
| 13 |
Hb_001184_090 |
0.0725141362 |
- |
- |
PREDICTED: TIP41-like protein [Jatropha curcas] |
| 14 |
Hb_000832_140 |
0.0726023888 |
- |
- |
actin depolymerizing factor 4 [Hevea brasiliensis] |
| 15 |
Hb_011689_080 |
0.0736998836 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 16 |
Hb_063716_050 |
0.0738061566 |
- |
- |
PREDICTED: uncharacterized protein LOC105634866 [Jatropha curcas] |
| 17 |
Hb_000107_050 |
0.0745829831 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 18 |
Hb_007576_040 |
0.0745880088 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 19 |
Hb_004800_230 |
0.0753807561 |
- |
- |
PREDICTED: MIP18 family protein At1g68310 [Jatropha curcas] |
| 20 |
Hb_002233_150 |
0.0755253375 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |