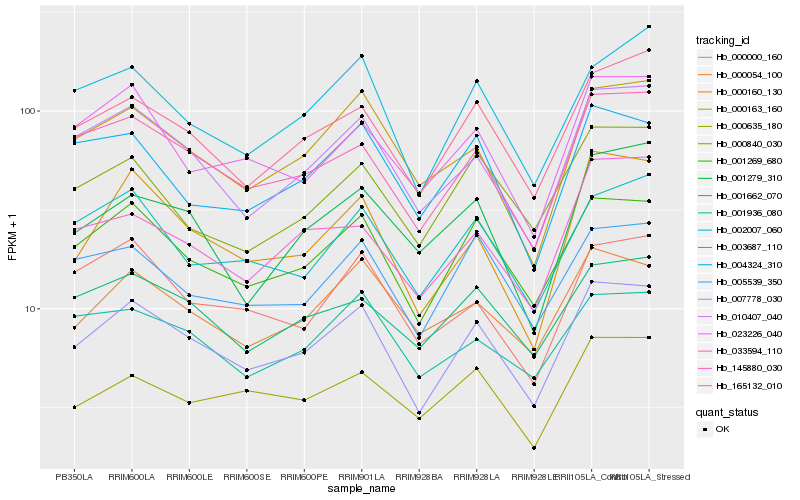
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007778_030 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001269_680 |
0.0495149281 |
- |
- |
PREDICTED: uncharacterized protein LOC105132670 isoform X2 [Populus euphratica] |
| 3 |
Hb_145880_030 |
0.0664445013 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 4 |
Hb_010407_040 |
0.0684497408 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 5 |
Hb_165132_010 |
0.0691635603 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 6 |
Hb_000163_160 |
0.0795782106 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 7 |
Hb_000160_130 |
0.0798054289 |
- |
- |
PREDICTED: uncharacterized protein LOC105648215 [Jatropha curcas] |
| 8 |
Hb_003687_110 |
0.081506375 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Malus domestica] |
| 9 |
Hb_002007_060 |
0.0825825166 |
- |
- |
hypothetical protein JCGZ_23323 [Jatropha curcas] |
| 10 |
Hb_005539_350 |
0.082929516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000054_100 |
0.0838642955 |
- |
- |
PREDICTED: metal tolerance protein C4 [Jatropha curcas] |
| 12 |
Hb_001662_070 |
0.0855673376 |
- |
- |
PREDICTED: thiol-disulfide oxidoreductase LTO1 [Jatropha curcas] |
| 13 |
Hb_023226_040 |
0.0882537484 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_004324_310 |
0.0912164459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000635_180 |
0.0913440584 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000000_160 |
0.0916905255 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 17 |
Hb_000840_030 |
0.0918260023 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 18 |
Hb_001936_080 |
0.093179907 |
- |
- |
PREDICTED: RNA-binding protein 48-like [Jatropha curcas] |
| 19 |
Hb_033594_110 |
0.0946739889 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 20 |
Hb_001279_310 |
0.0976889173 |
- |
- |
hypothetical protein JCGZ_10212 [Jatropha curcas] |