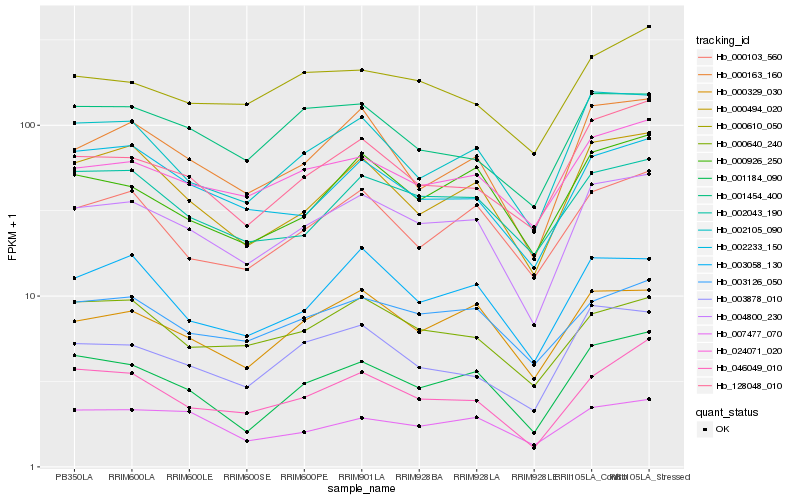
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_230 |
0.0 |
- |
- |
PREDICTED: MIP18 family protein At1g68310 [Jatropha curcas] |
| 2 |
Hb_000329_030 |
0.0703976463 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_024071_020 |
0.0724085095 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 4 |
Hb_000163_160 |
0.0725610394 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 5 |
Hb_003126_050 |
0.0746902827 |
- |
- |
PREDICTED: cell cycle checkpoint protein RAD17 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000494_020 |
0.0753807561 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 5A [Jatropha curcas] |
| 7 |
Hb_001184_090 |
0.0760505596 |
- |
- |
PREDICTED: TIP41-like protein [Jatropha curcas] |
| 8 |
Hb_002105_090 |
0.0782598966 |
- |
- |
Rab4 [Hevea brasiliensis] |
| 9 |
Hb_003058_130 |
0.0823926108 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A isoform X1 [Jatropha curcas] |
| 10 |
Hb_002043_190 |
0.083036127 |
- |
- |
rwd domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003878_010 |
0.0838420409 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 12 |
Hb_128048_010 |
0.0839746608 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 13 |
Hb_002233_150 |
0.0846496888 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 14 |
Hb_000926_250 |
0.0846732643 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 15 |
Hb_000103_560 |
0.0853747592 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 16 |
Hb_000640_240 |
0.0859263547 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 17 |
Hb_046049_010 |
0.0870775255 |
- |
- |
PREDICTED: far upstream element-binding protein 2 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000610_050 |
0.0875295918 |
- |
- |
skp1, putative [Ricinus communis] |
| 19 |
Hb_007477_070 |
0.0877276231 |
- |
- |
hypothetical protein JCGZ_17593 [Jatropha curcas] |
| 20 |
Hb_001454_400 |
0.0880245246 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |