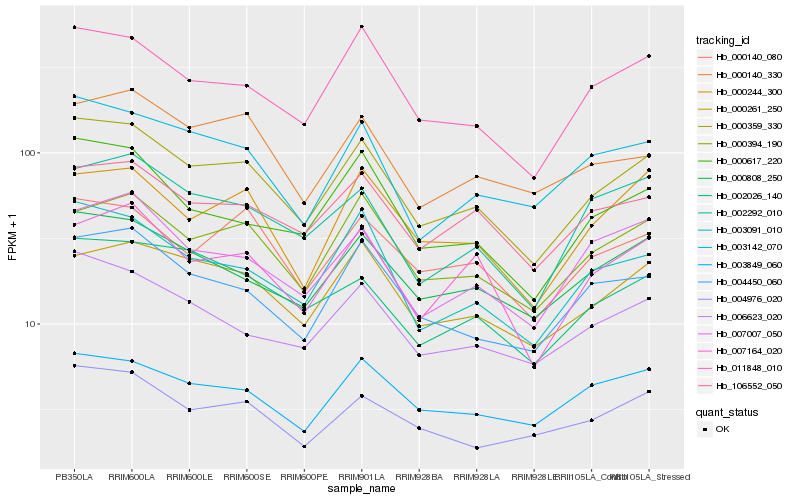
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011848_010 |
0.0667342491 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 3 |
Hb_003091_010 |
0.0693284501 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 4 |
Hb_000808_250 |
0.0706652603 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 5 |
Hb_000394_190 |
0.0738470551 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_006623_020 |
0.0760311995 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004450_060 |
0.076982083 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003142_070 |
0.0772918564 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 9 |
Hb_000261_250 |
0.0774422733 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 10 |
Hb_000617_220 |
0.0799406464 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 11 |
Hb_106552_050 |
0.0811985998 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 12 |
Hb_000140_080 |
0.0819233777 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004976_020 |
0.0833640179 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 14 |
Hb_007164_020 |
0.0846729576 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 15 |
Hb_007007_050 |
0.0855655 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 16 |
Hb_003849_060 |
0.0856965244 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000244_300 |
0.086140543 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002026_140 |
0.0879769364 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 19 |
Hb_002292_010 |
0.0880640643 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 20 |
Hb_000140_330 |
0.0904887494 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |