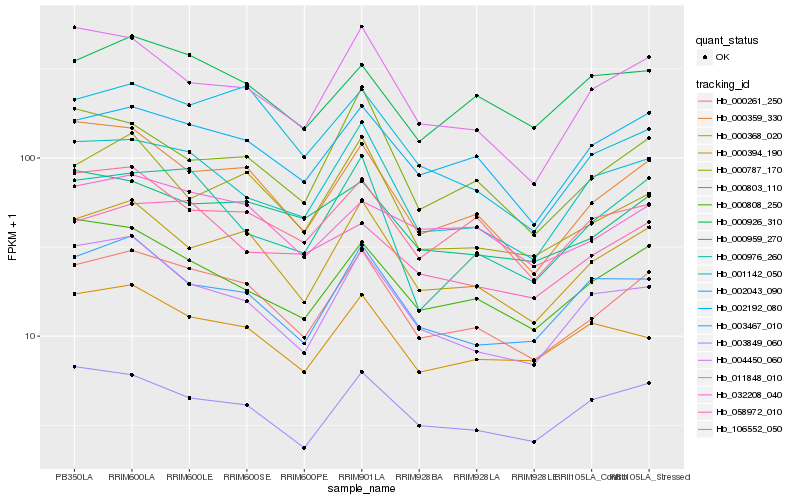
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_250 |
0.0 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 2 |
Hb_000394_190 |
0.0547159001 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_002192_080 |
0.0652085271 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 4 |
Hb_001142_050 |
0.0734808557 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 5 |
Hb_000359_330 |
0.0774422733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003849_060 |
0.0816782405 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 7 |
Hb_106552_050 |
0.083869415 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 8 |
Hb_000959_270 |
0.0850227214 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 9 |
Hb_000808_250 |
0.0853885544 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 10 |
Hb_003467_010 |
0.0880435005 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 11 |
Hb_000787_170 |
0.088228304 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 12 |
Hb_032208_040 |
0.0884214096 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 13 |
Hb_011848_010 |
0.0885110406 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 14 |
Hb_002043_090 |
0.0900997519 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 15 |
Hb_004450_060 |
0.0905727597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000976_260 |
0.0917326132 |
- |
- |
- |
| 17 |
Hb_000803_110 |
0.0922541482 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 18 |
Hb_058972_010 |
0.0922670879 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000368_020 |
0.0928472122 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000926_310 |
0.0931280096 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |