| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
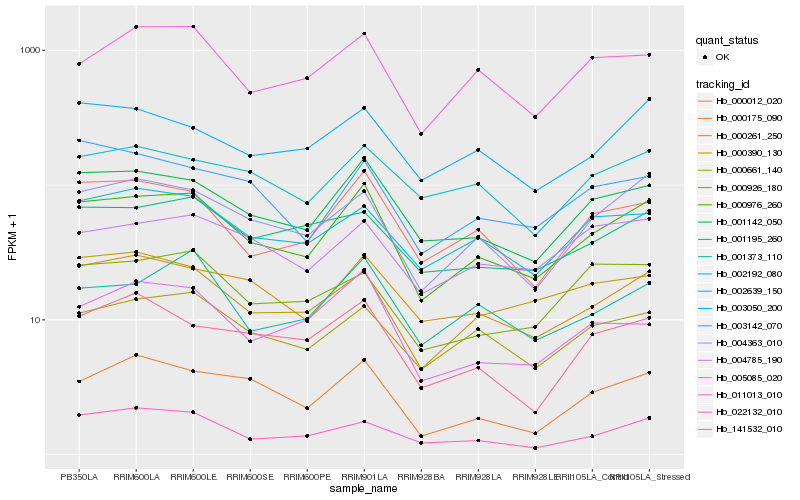
Hb_000976_260 |
0.0 |
- |
- |
- |
| 2 |
Hb_001142_050 |
0.0683904264 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 3 |
Hb_000012_020 |
0.0853580042 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 4 |
Hb_004363_010 |
0.0860752842 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 5 |
Hb_000261_250 |
0.0917326132 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 6 |
Hb_000661_140 |
0.0958447044 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002639_150 |
0.1018351873 |
- |
- |
- |
| 8 |
Hb_001373_110 |
0.1034041477 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 9 |
Hb_000175_090 |
0.1046575649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011013_010 |
0.1059465837 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_003050_200 |
0.107674871 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 12 |
Hb_022132_010 |
0.1094086741 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_005085_020 |
0.1106218986 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 14 |
Hb_001195_260 |
0.1117654826 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 15 |
Hb_004785_190 |
0.1133296732 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 16 |
Hb_002192_080 |
0.113515948 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 17 |
Hb_000390_130 |
0.1137394818 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 18 |
Hb_003142_070 |
0.1139498526 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 19 |
Hb_000926_180 |
0.1144104339 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 20 |
Hb_141532_010 |
0.1150381619 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |