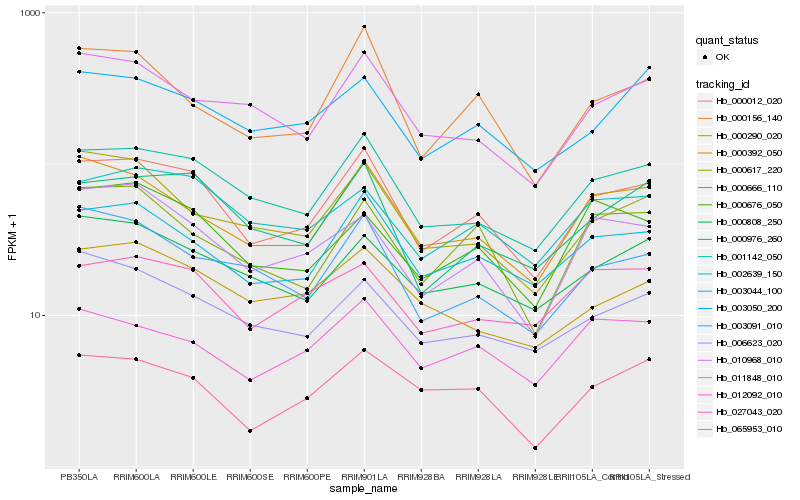
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_020 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 2 |
Hb_001142_050 |
0.0656389253 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 3 |
Hb_003044_100 |
0.0758867191 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 4 |
Hb_000976_260 |
0.0853580042 |
- |
- |
- |
| 5 |
Hb_000290_020 |
0.0879257547 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 6 |
Hb_002639_150 |
0.0962321321 |
- |
- |
- |
| 7 |
Hb_000156_140 |
0.0998364186 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 8 |
Hb_000808_250 |
0.101662382 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 9 |
Hb_003091_010 |
0.1022314549 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 10 |
Hb_011848_010 |
0.1024580332 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 11 |
Hb_010968_010 |
0.1032157745 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 12 |
Hb_027043_020 |
0.1061590255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012092_010 |
0.1067844336 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 14 |
Hb_006623_020 |
0.1072952173 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000666_110 |
0.1076487649 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 16 |
Hb_000617_220 |
0.1079214225 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 17 |
Hb_000676_050 |
0.1084539661 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 18 |
Hb_000392_050 |
0.1087671177 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 19 |
Hb_065953_010 |
0.1088074521 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 20 |
Hb_003050_200 |
0.1098289738 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |