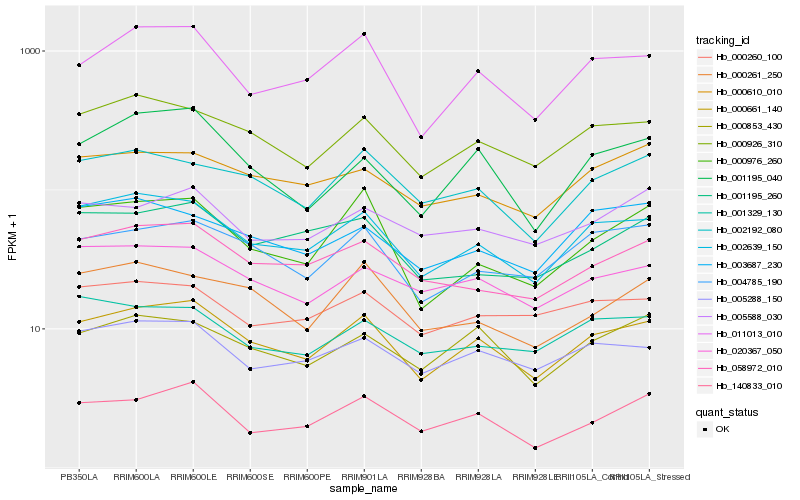
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002639_150 |
0.0519374371 |
- |
- |
- |
| 3 |
Hb_000926_310 |
0.0633286625 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 4 |
Hb_011013_010 |
0.0703024416 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 5 |
Hb_004785_190 |
0.0789610916 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 6 |
Hb_005288_150 |
0.0804487165 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_020367_050 |
0.0827130666 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_002192_080 |
0.0829561378 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 9 |
Hb_058972_010 |
0.083333483 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000853_430 |
0.083578067 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 11 |
Hb_140833_010 |
0.0892945629 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 12 |
Hb_001195_040 |
0.0894919612 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 13 |
Hb_000610_010 |
0.0925676905 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_000976_260 |
0.0958447044 |
- |
- |
- |
| 15 |
Hb_000261_250 |
0.0961939173 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 16 |
Hb_001195_260 |
0.0988260643 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 17 |
Hb_000260_100 |
0.0992368129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003687_230 |
0.1001883275 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 19 |
Hb_005588_030 |
0.1009476607 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 20 |
Hb_001329_130 |
0.1010139483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |