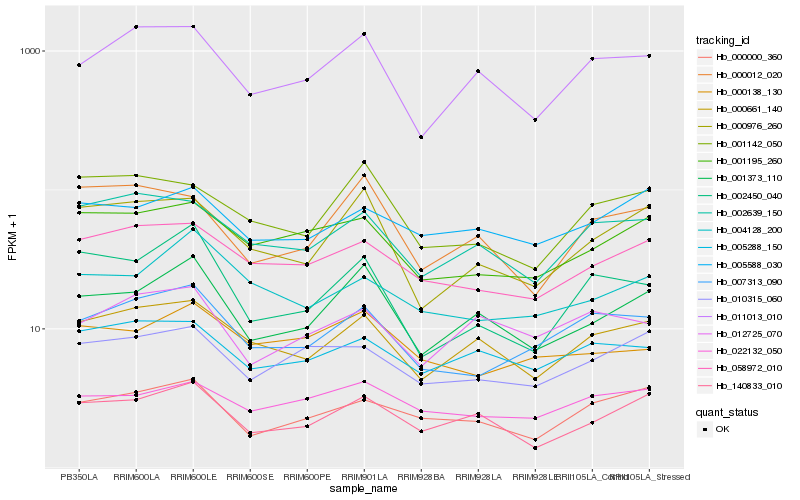
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001373_110 |
0.0 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 2 |
Hb_140833_010 |
0.0810715545 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 3 |
Hb_011013_010 |
0.1020139788 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 4 |
Hb_000661_140 |
0.1030844434 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000976_260 |
0.1034041477 |
- |
- |
- |
| 6 |
Hb_000012_020 |
0.1231847757 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 7 |
Hb_002450_040 |
0.1288673952 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 8 |
Hb_000000_360 |
0.1298667354 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 9 |
Hb_001195_260 |
0.1301813583 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 10 |
Hb_001142_050 |
0.1321619493 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 11 |
Hb_002639_150 |
0.1331221374 |
- |
- |
- |
| 12 |
Hb_005588_030 |
0.1335696342 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 13 |
Hb_058972_010 |
0.134079905 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007313_090 |
0.1358761471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012725_070 |
0.1364417403 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004128_200 |
0.1372335392 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 17 |
Hb_000138_130 |
0.137802894 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 18 |
Hb_010315_060 |
0.1382141352 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 19 |
Hb_005288_150 |
0.138761228 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_022132_050 |
0.1389137429 |
- |
- |
GTP binding protein, putative [Ricinus communis] |