| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
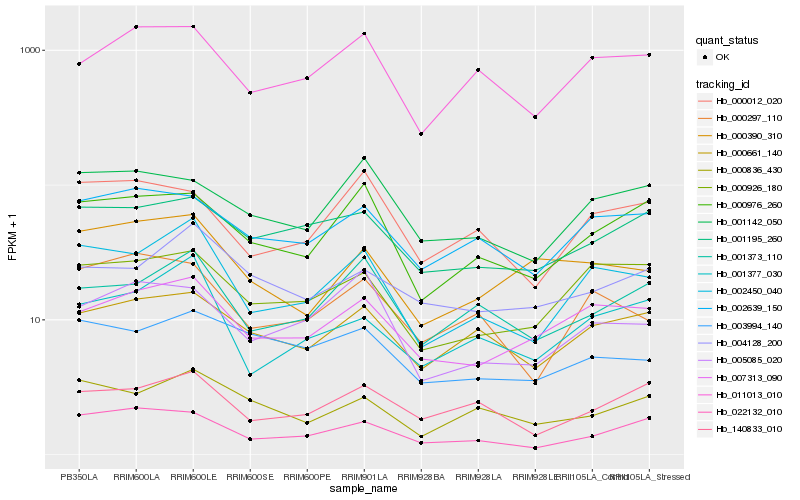
Hb_002450_040 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 2 |
Hb_001377_030 |
0.1167515731 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 3 |
Hb_000926_180 |
0.120423864 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 4 |
Hb_007313_090 |
0.1252819536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001373_110 |
0.1288673952 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 6 |
Hb_000297_110 |
0.1302244884 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 7 |
Hb_000976_260 |
0.1317043079 |
- |
- |
- |
| 8 |
Hb_011013_010 |
0.1377742205 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 9 |
Hb_002639_150 |
0.1384828009 |
- |
- |
- |
| 10 |
Hb_000661_140 |
0.1386584166 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 11 |
Hb_022132_010 |
0.1406443966 |
- |
- |
unknown [Medicago truncatula] |
| 12 |
Hb_000012_020 |
0.1420421926 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 13 |
Hb_000836_430 |
0.1429342464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_140833_010 |
0.1457765566 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 15 |
Hb_005085_020 |
0.1476186448 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 16 |
Hb_001142_050 |
0.1489958402 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 17 |
Hb_001195_260 |
0.1499352907 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 18 |
Hb_000390_310 |
0.1515155506 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003994_140 |
0.1518880166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004128_200 |
0.1522285308 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |