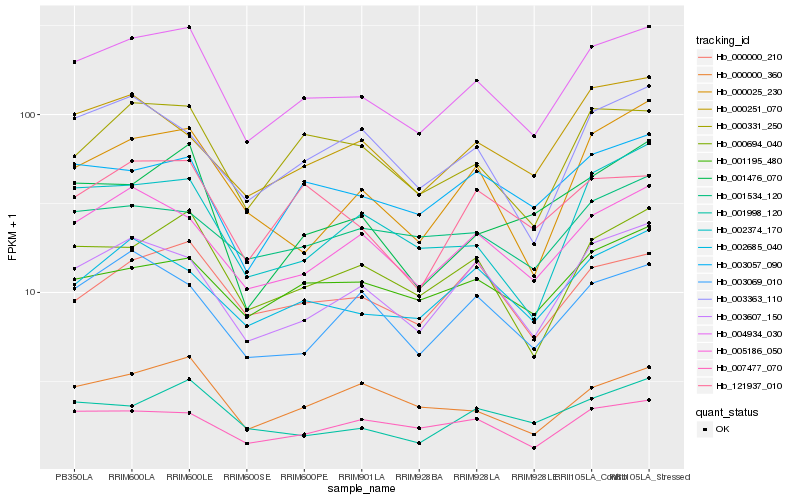
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004934_030 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 2 |
Hb_000694_040 |
0.0779689781 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_003607_150 |
0.082075516 |
- |
- |
PREDICTED: thioredoxin-like protein 4B [Jatropha curcas] |
| 4 |
Hb_005186_050 |
0.0830239889 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 5 |
Hb_000331_250 |
0.0892506948 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 6 |
Hb_000000_210 |
0.0936574948 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001998_120 |
0.0946962845 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 8 |
Hb_002685_040 |
0.0956916047 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 9 |
Hb_000000_360 |
0.0958377368 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 10 |
Hb_003057_090 |
0.0990861686 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 11 |
Hb_001195_480 |
0.1019809464 |
- |
- |
cop9 complex subunit, putative [Ricinus communis] |
| 12 |
Hb_121937_010 |
0.1022774302 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 13 |
Hb_007477_070 |
0.1026138887 |
- |
- |
hypothetical protein JCGZ_17593 [Jatropha curcas] |
| 14 |
Hb_003069_010 |
0.1046872783 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 15 |
Hb_002374_170 |
0.1073193767 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 16 |
Hb_003363_110 |
0.1076146361 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001534_120 |
0.1080925707 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 18 |
Hb_000251_070 |
0.1084682784 |
- |
- |
hypothetical protein L484_010641 [Morus notabilis] |
| 19 |
Hb_001476_070 |
0.1085330084 |
- |
- |
MinE protein [Manihot esculenta] |
| 20 |
Hb_000025_230 |
0.1088036762 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |