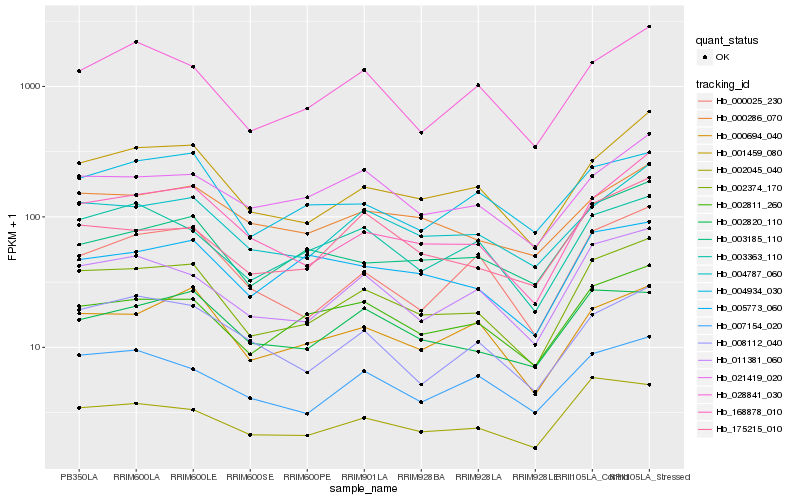
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_170 |
0.0 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 2 |
Hb_011381_060 |
0.0755902263 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 3 |
Hb_001459_080 |
0.0765747069 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 4 |
Hb_004787_060 |
0.0811808833 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 5 |
Hb_168878_010 |
0.0815947204 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 6 |
Hb_002811_260 |
0.0928750769 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 7 |
Hb_028841_030 |
0.0949341308 |
- |
- |
- |
| 8 |
Hb_000694_040 |
0.0953918715 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_002045_040 |
0.0957337668 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_003363_110 |
0.0964276254 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007154_020 |
0.0968247261 |
- |
- |
- |
| 12 |
Hb_175215_010 |
0.0989157867 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 13 |
Hb_000025_230 |
0.0989998285 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |
| 14 |
Hb_000286_070 |
0.1016885572 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 15 |
Hb_008112_040 |
0.104854194 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 16 |
Hb_021419_020 |
0.1065439454 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 17 |
Hb_004934_030 |
0.1073193767 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 18 |
Hb_003185_110 |
0.1075917923 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 7-2 [Nelumbo nucifera] |
| 19 |
Hb_002820_110 |
0.1076246065 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 20 |
Hb_005773_060 |
0.1096259168 |
- |
- |
defender against cell death, putative [Ricinus communis] |