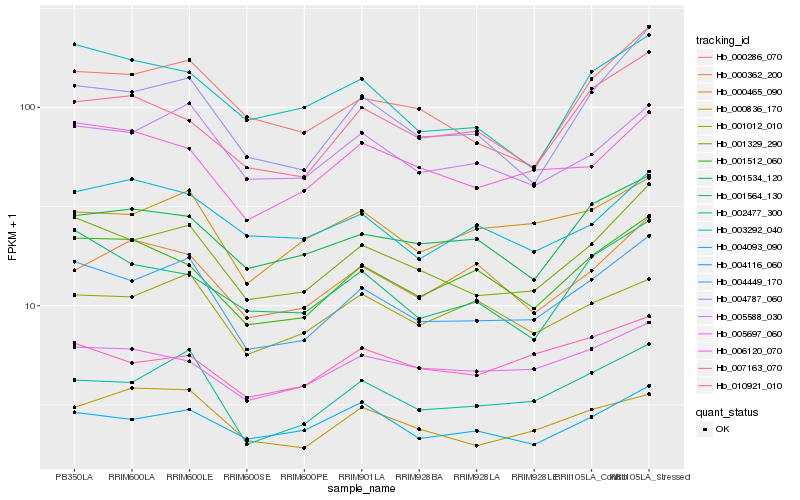
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001329_290 |
0.0436292093 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 3 |
Hb_002477_300 |
0.0544723817 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_005588_030 |
0.0739375471 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 5 |
Hb_004787_060 |
0.0740106309 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 6 |
Hb_001512_060 |
0.0772107306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000286_070 |
0.0802564039 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 8 |
Hb_000465_090 |
0.0819241458 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 9 |
Hb_006120_070 |
0.0831116188 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 10 |
Hb_001534_120 |
0.0842707791 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 11 |
Hb_001564_130 |
0.0877701672 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_010921_010 |
0.0880992937 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 13 |
Hb_000836_170 |
0.0887375499 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 14 |
Hb_005697_060 |
0.0910970841 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_004116_060 |
0.0914118157 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 16 |
Hb_001012_010 |
0.0930794791 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 17 |
Hb_003292_040 |
0.0937941234 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 18 |
Hb_007163_070 |
0.0956866737 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000362_200 |
0.095895574 |
- |
- |
exonuclease family protein [Populus trichocarpa] |
| 20 |
Hb_004093_090 |
0.0979944983 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |