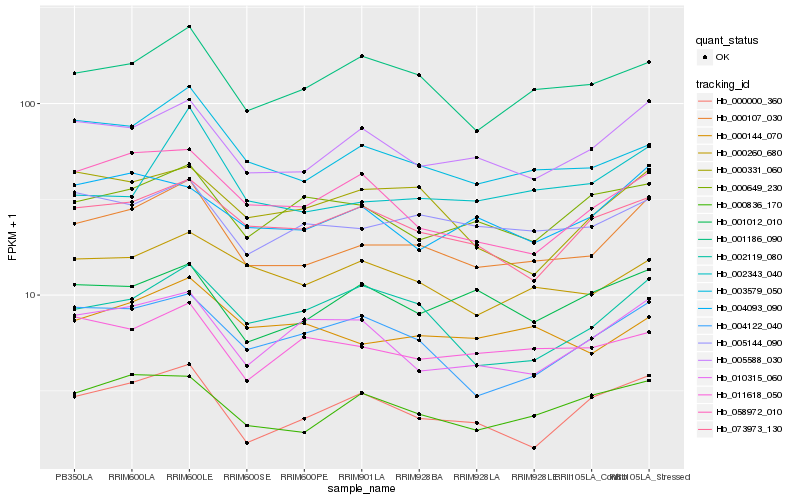
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005588_030 |
0.0678135 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 3 |
Hb_003579_050 |
0.0774789614 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 4 |
Hb_005144_090 |
0.085184908 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 5 |
Hb_000836_170 |
0.0854603762 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 6 |
Hb_001186_090 |
0.0877057405 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 7 |
Hb_073973_130 |
0.0930704903 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000260_680 |
0.0947036592 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 9 |
Hb_000144_070 |
0.0961046392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_058972_010 |
0.0982482454 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004093_090 |
0.0983218151 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 12 |
Hb_011618_050 |
0.0987888169 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 13 |
Hb_004122_040 |
0.0988655828 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 14 |
Hb_001012_010 |
0.0999533613 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 15 |
Hb_002343_040 |
0.100370765 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000649_230 |
0.1012931028 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 17 |
Hb_010315_060 |
0.1022537213 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 18 |
Hb_002119_080 |
0.1026847127 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000331_060 |
0.1031005837 |
- |
- |
PREDICTED: 60S ribosomal protein L17-2-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000000_360 |
0.103403219 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |