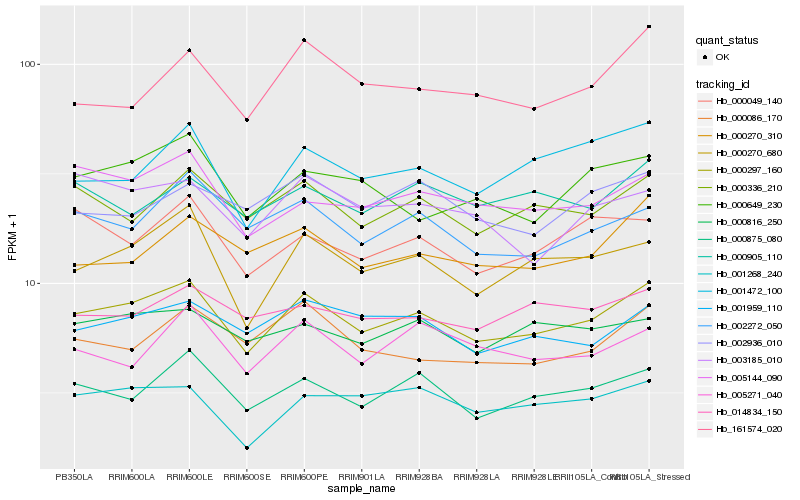
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000297_160 |
0.0 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 2 |
Hb_000875_080 |
0.0637406889 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_000270_680 |
0.0638435275 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 4 |
Hb_000336_210 |
0.067808483 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001959_110 |
0.0709674027 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005144_090 |
0.0713533406 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 7 |
Hb_002272_050 |
0.0718961273 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 8 |
Hb_001268_240 |
0.0750828793 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000816_250 |
0.0761724705 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001472_100 |
0.0766502836 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 11 |
Hb_000270_310 |
0.076685202 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 12 |
Hb_000049_140 |
0.0777156689 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000649_230 |
0.0778178334 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 14 |
Hb_000086_170 |
0.0785249761 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 15 |
Hb_014834_150 |
0.0793510775 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000905_110 |
0.0806520215 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 17 |
Hb_002936_010 |
0.0813513197 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 18 |
Hb_161574_020 |
0.0816974609 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_005271_040 |
0.0819910196 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 20 |
Hb_003185_010 |
0.0820597119 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |