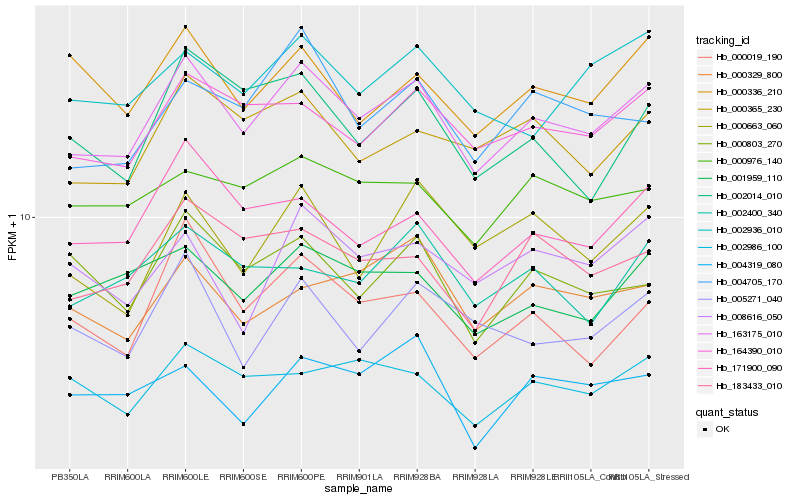
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163175_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 2 |
Hb_164390_010 |
0.0537874989 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 3 |
Hb_171900_090 |
0.0597855866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000365_230 |
0.061884671 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 5 |
Hb_001959_110 |
0.0629477166 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 6 |
Hb_183433_010 |
0.0629908715 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 7 |
Hb_004705_170 |
0.0660708586 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 8 |
Hb_000976_140 |
0.0666887543 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000019_190 |
0.0678499857 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 10 |
Hb_002936_010 |
0.0681664673 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 11 |
Hb_002986_100 |
0.068477665 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 12 |
Hb_002400_340 |
0.0692981175 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 13 |
Hb_005271_040 |
0.0696442958 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 14 |
Hb_000329_800 |
0.0697814393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000663_060 |
0.0700327559 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 16 |
Hb_000803_270 |
0.0708806313 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 17 |
Hb_000336_210 |
0.0711063023 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002014_010 |
0.0712818789 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_008616_050 |
0.0714771474 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 20 |
Hb_004319_080 |
0.071707631 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |