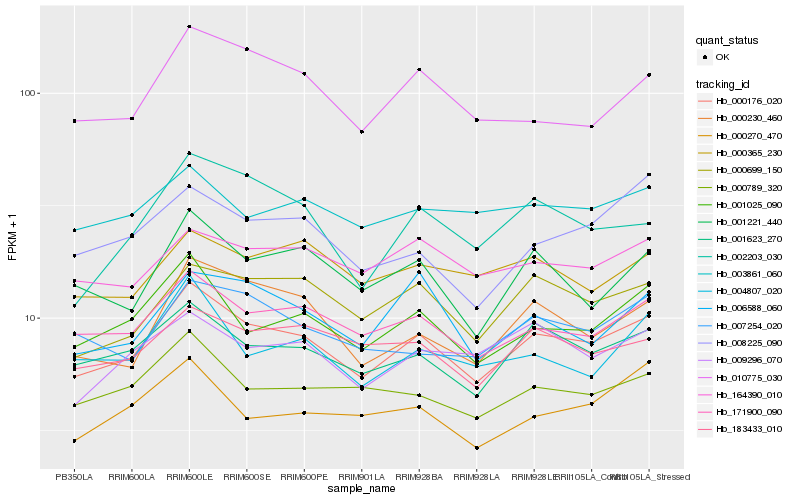
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000176_020 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 2 |
Hb_171900_090 |
0.0589447763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001623_270 |
0.0609926471 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 4 |
Hb_001025_090 |
0.062611026 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 5 |
Hb_000230_460 |
0.0725363621 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 6 |
Hb_000699_150 |
0.0729066568 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_000789_320 |
0.0751223317 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001221_440 |
0.0777639272 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_183433_010 |
0.0779140549 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_007254_020 |
0.0795820535 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 11 |
Hb_004807_020 |
0.0810019637 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 12 |
Hb_010775_030 |
0.0813239313 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 13 |
Hb_006588_060 |
0.0819465672 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 14 |
Hb_164390_010 |
0.0822447476 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 15 |
Hb_009296_070 |
0.0824765433 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003861_060 |
0.0835391908 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 17 |
Hb_002203_030 |
0.0860140864 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_000365_230 |
0.0864938602 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 19 |
Hb_000270_470 |
0.0868728311 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: pentatricopeptide repeat-containing protein At1g74900, mitochondrial [Jatropha curcas] |
| 20 |
Hb_008225_090 |
0.0870113507 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |