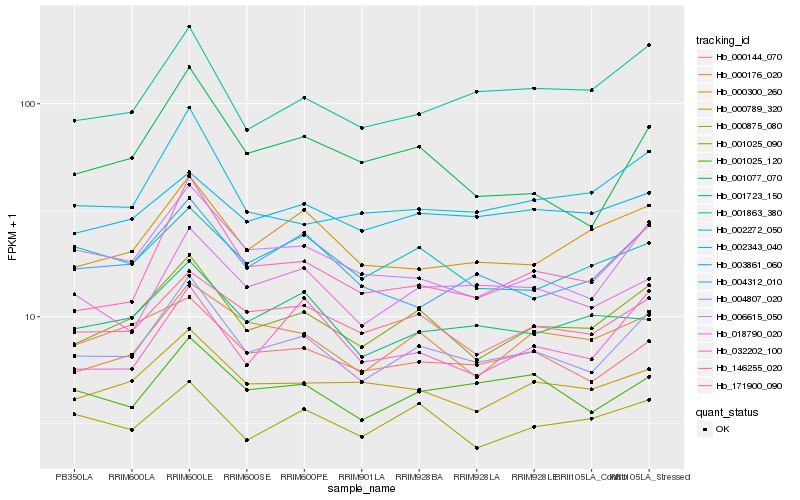
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004807_020 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 2 |
Hb_001025_090 |
0.0549699064 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 3 |
Hb_006615_050 |
0.0642492156 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 4 |
Hb_001025_120 |
0.0717248188 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_171900_090 |
0.0724413675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_146255_020 |
0.0736122102 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 7 |
Hb_018790_020 |
0.0758918468 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_002343_040 |
0.0797679689 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001863_380 |
0.0801422228 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 10 |
Hb_000176_020 |
0.0810019637 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 11 |
Hb_000789_320 |
0.0831720839 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004312_010 |
0.083684666 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000300_260 |
0.0844587003 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 14 |
Hb_001077_070 |
0.0846208246 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 15 |
Hb_002272_050 |
0.0891185211 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 16 |
Hb_001723_150 |
0.0901142275 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000144_070 |
0.0901905456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003861_060 |
0.090506003 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 19 |
Hb_000875_080 |
0.0910822792 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_032202_100 |
0.0919705117 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |