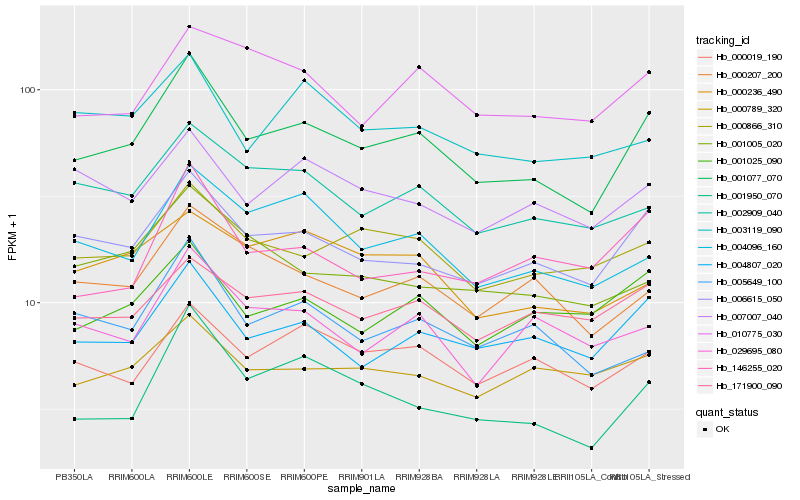
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001077_070 |
0.0 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 2 |
Hb_006615_050 |
0.0738378516 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 3 |
Hb_004807_020 |
0.0846208246 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 4 |
Hb_001950_070 |
0.0942888373 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 5 |
Hb_001025_090 |
0.0967346652 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 6 |
Hb_000866_310 |
0.0969035503 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 7 |
Hb_002909_040 |
0.0989288881 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 8 |
Hb_000019_190 |
0.1008051631 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 9 |
Hb_004096_160 |
0.1013207713 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 10 |
Hb_000207_200 |
0.1013672986 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_171900_090 |
0.1030345338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_146255_020 |
0.1058440139 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 13 |
Hb_007007_040 |
0.1059084894 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 14 |
Hb_005649_100 |
0.1060352773 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 15 |
Hb_010775_030 |
0.1065366796 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 16 |
Hb_001005_020 |
0.1083259117 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 17 |
Hb_000789_320 |
0.1084592809 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_029695_080 |
0.1086509415 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 19 |
Hb_000236_490 |
0.1094093266 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 20 |
Hb_003119_090 |
0.1105921161 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |