| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
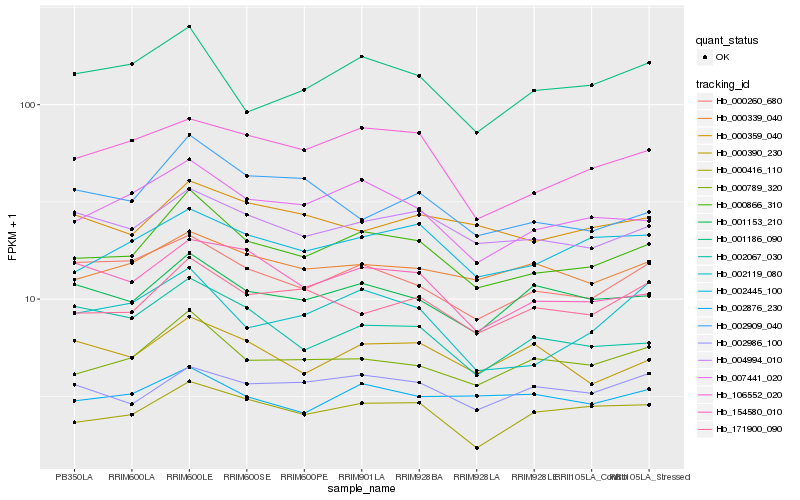
Hb_000866_310 |
0.0 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 2 |
Hb_007441_020 |
0.0625589318 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 3 |
Hb_000789_320 |
0.0644537456 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004994_010 |
0.0738293497 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_000260_680 |
0.0744136459 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 6 |
Hb_000416_110 |
0.0744192262 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002445_100 |
0.0757218304 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001153_210 |
0.0770560075 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 9 |
Hb_171900_090 |
0.0770897062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_106552_020 |
0.0782496649 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 11 |
Hb_154580_010 |
0.0792562059 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 12 |
Hb_001186_090 |
0.081516203 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 13 |
Hb_000339_040 |
0.0819589049 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_002067_030 |
0.086622146 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000359_040 |
0.0887972816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002876_230 |
0.0894460287 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 17 |
Hb_000390_230 |
0.0897922282 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 18 |
Hb_002986_100 |
0.0898993934 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 19 |
Hb_002909_040 |
0.0903438641 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 20 |
Hb_002119_080 |
0.0907525249 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |