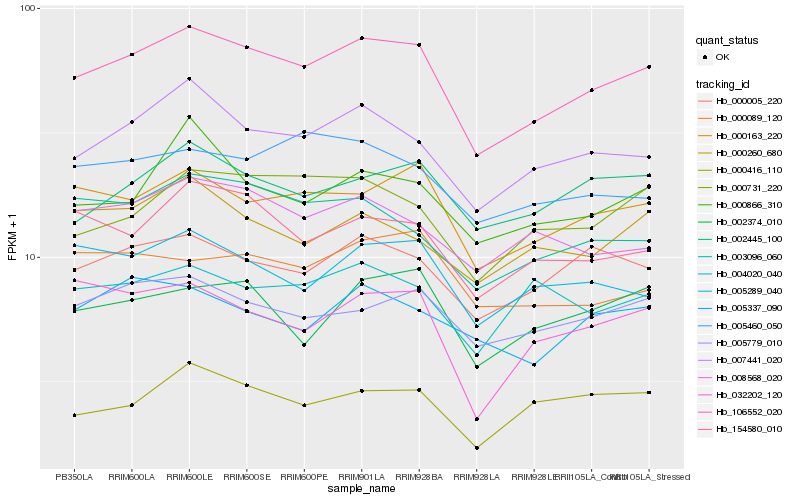
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106552_020 |
0.0 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007441_020 |
0.060043012 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 3 |
Hb_000163_220 |
0.0618830741 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 4 |
Hb_000005_220 |
0.0690170161 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 5 |
Hb_005779_010 |
0.071346021 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_154580_010 |
0.0727248075 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 7 |
Hb_000731_220 |
0.0738991678 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 8 |
Hb_002445_100 |
0.075052124 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003096_060 |
0.0765862482 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 10 |
Hb_005289_040 |
0.0769403447 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 11 |
Hb_000416_110 |
0.0780149357 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002374_010 |
0.0781185193 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000866_310 |
0.0782496649 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 14 |
Hb_008568_020 |
0.079037439 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 15 |
Hb_005460_050 |
0.0815361457 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 16 |
Hb_000089_120 |
0.0816050113 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000260_680 |
0.0821807154 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 18 |
Hb_005337_090 |
0.083084082 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 19 |
Hb_004020_040 |
0.0841356249 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 20 |
Hb_032202_120 |
0.0851171584 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |